Figure 3.
Tridimensional Architecture of miR-206/133b Genomic Locus
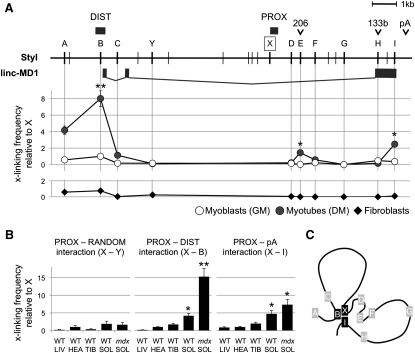
(A) Upper panel shows StyI restriction sites (vertical lines), regulatory regions (DIST, PROX, pA), pre-miRNAs (206, 133b), and linc-MD1. Capital letters (A–I, X, and Y) indicate the position of StyI sites analyzed in 3C experiments. A–I sites identify the same regions analyzed in ChIP experiments in Figure 2C. A common reverse primer identifying the proximal region (X) was used in combination with different reverse primers. Lower panel shows 3C analysis performed on chromatin extracted from C2 myoblasts in GM and DM conditions and from fibroblast cell cultures. Crosslinking frequencies relative to X region were measured both by semiquantitative PCR (data not shown) and qRT-PCR. Data from multiple experiments were normalized for primer amplification efficiency and reported with respect to X-B interaction in GM set to a value of 1. Statistical significance was calculated with respect to GM conditions.
(B) The same 3C analysis was performed in liver (LIV), heart (HEA), tibialis anterior (TIB), soleus (SOL) of wild-type (WT), and mdx animals. For each set of interactions analyzed, crosslinking frequencies relative to X region derived from multiple experiments were reported with respect to WT HEA interaction set to a value of 1. Statistical significance was calculated with respect to WT HEA tissue. Data are shown as mean ± SD. One asterisk indicates p < 0.05; two asterisks: p < 0.01.
(C) Schematic representation of the interactions detected upon induction of differentiation.