Figure S1.
Related to Figure 1
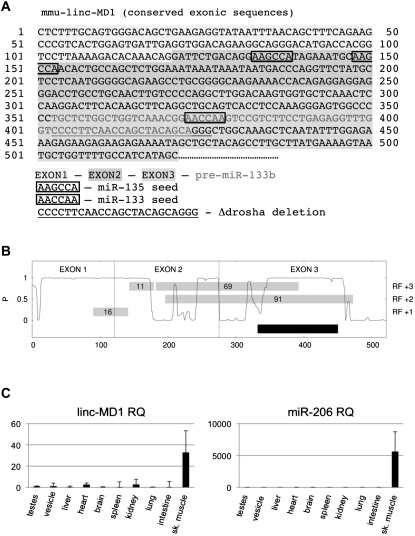
(A) Murine linc-MD1 with exons identification, miRNA binding sites and sequence deleted in Δdrosha constructs. The nucleotide sequence refers to the portion of the transcript which is conserved between human and mouse. Two 3′-end isoforms mapping at two pA-sites have been mapped in a non conserved region at 1060 and 1450 nt downstream the indicated sequence (Figure 1B).
(B) Conservation pattern of the linc-MD1 transcript expressed as probability of negative selection taken from the Mammal Cons track. RF+1, +2 and +3 refer to the reading frames, gray boxes indicate the position of ORFs in number of triplets. The mature transcript contains four open reading frames (defined as any sequence starting with AUG and containing at least 10 non terminator codons) ranging in size from 11 to 91 triplets. None of the AUG shows a Kozak consensus (Kozak 1989), nor are the sequences more or less conserved than the surrounding regions.
(C) linc-MD1 and miR-206 relative quantifications in the indicated tisses. Values are normalized for HPRT or U6 snRNA respectively and shown with respect to testes sample set to a value of 1.