Abstract
Lysophosphatidic acid (LPA) is a bioactive mediator and induces several biological effects, including cell proliferation, migration, morphogenesis and differentiation. LPA interacts with at least six G protein-coupled receptors (GPCRs), including LPA receptor-1 (LPA1), LPA2, LPA3, LPA4, LPA5 and LPA6. These receptors show different biological functions through the binding of LPA, depending on the type of cells. In human malignancies, a high level of LPA production was found in plasma and ascites in ovarian cancer cases. Moreover, aberrant expression levels of LPA receptor genes were detected in some cancer cells. Therefore, it is suggested that LPA receptors may be involved in the pathogenesis of tumor cells as well as LPA per se. Recently, we have reported that alterations of LPA receptor genes also occur in rodent tumors. In this review, we summarize the recent evidence in the investigations of LPA receptor alterations in rodent tumors by experimental models.
Keywords: LPA, LPA receptor, mutation, DNA methylation, rodent
Introduction
Lysophosphatidic acid (LPA) is a simple bioactive phospholipid consisting of a phosphate, a fatty acid and a glycerol1,2. It is present in all mammalian cells and tissues and is detected in serum, plasma, saliva and activated platelets1–3. LPA induces several cellular responses, such as cell proliferation, differentiation, morphogenesis, cell migration, platelet aggregation, secretion of cytokine and chemokine and protection from apoptosis1–3. Moreover, it is suggested that LPA may be also involved in the development of human diseases, including cancers2,4. In fact, a high level of LPA production was found in plasma and ascites from patients with ovarian cancers, and the plasma levels of LPA were also increased in chronic hepatitis C in association with liver fibrosis1,5. LPA enhanced malignant abilities of tumor cells, including cell growth, migration, invasion, production of angiogenic factors and tumorigenicity2,4.
LPA interacts with G protein-coupled receptors (GPCRs) to induce various responses. So far, at least six LPA receptors have been identified1–4. Previously, aberrant expressions of LPA receptor genes have been also detected in human malignancies, such as ovary, colon and thyroid tumors6–9. Moreover, the exogenous LPAR2 or LPAR3-expressing ovarian cancer cells increased malignant properties, such as cell migration, invasion and tumorigenicity10. Therefore, it has been demonstrated that LPA receptors may be involved in the acquisition of malignant potency of tumor cells as well as LPA per se. In our energetic studies, we have indicated that mutations and aberrant expressions due to DNA methylation of LPA receptor genes occur in rodent tumors induced by experimental models. Here, we review the current knowledge about LPA receptor gene alterations in rodent tumors and suggest an involvement of the LPA signaling pathway during carcinogenesis of rodents.
LPA Receptors
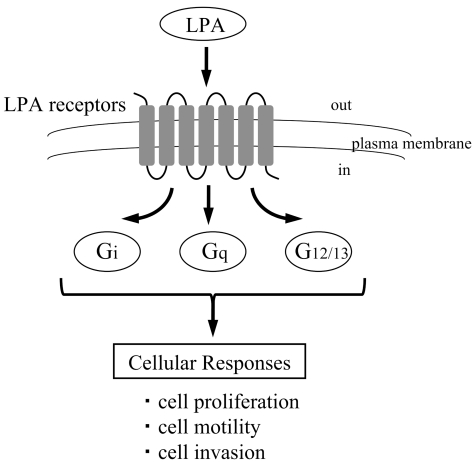
LPA acts as a biological mediator binding with LPA specific G protein-coupled transmembrane receptors, LPA receptors. So far, at least six LPA receptors have been identified, including LPA1/EDG2, LPA2/EDG4, LPA3/EDG7, LPA4/P2Y9/GPR23, LPA5/GPR92 and LPA6/P2Y51–4. Additionally, GPR87 has been proposed as a candidate for a new LPA receptor11. LPA receptors couple with individual sets of G proteins (Gi, Gq and G12/13) and mediate various biological responses to LPA1–4. The expression patterns of LPA receptor genes are dependent on the type of cells1. While LPA1 is ubiquitously expressed in normal tissues, the expressions of other LPA receptors are relatively restricted, suggesting that these receptors have different biological functions1. For example, both LPA1 and LPA2 can couple with Gi, Gq and G12/13, and stimulates cell proliferation, phospholipase C activation, intracellular calcium mobilization and adenylyl cyclase inhibition12. However, some different phenotypes of LPA1 and LPA2 have also been found in knockout mice, indicating that LPA1 and LPA2, at least in part, possess distinct functions13. On the other hand, LPA3 couples with Gi and Gq, but not G12/13. In recent studies, LPA6/P2Y5 has been identified as a causative gene of hair loss14,15 (Fig.1).
Fig. 1.
Cellular responses of LPA receptors through the binding of LPA.
Genetic Alterations
Mutations for LPA receptor genes were found in human colon cancer and osteosarcoma cells16,17. In colon cancer cells, 2 out of 6 cells showed LPAR2 and/or LPAR4 gene mutations16. LPAR1 and LPAR3 gene mutations occurred in one out of two osteosarcoma cells17. In rodent tumors, different patterns and frequencies of LPA receptor gene mutations were also observed.
Mutations of LPA receptor genes in rodent tumors
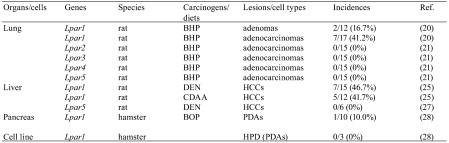
Lung: A mutation analysis for LPA receptor genes was performed using rat lung lesions induced by N-nitrosobis(2-hydroxypropyl)amine (BHP)18,19. This rat lung carcinogenesis model can obtain high yields of adenomatous lesions from preneoplastic lesions to carcinomas by continuous oral administration of BHP. Although no mutation of the Lpar1 gene was found in 15 hyperplasias, two out of 12 adenomas (16.7%) showed a TGC to CGC (Cys to Arg) transition at codon 24 and a TAT to TAC (Tyr to Tyr) transition at codon 292, respectively. Moreover, the Lpar1 gene mutations were detected in 7 out of 17 adenocarcinomas (41.2%). In the 7 adenocarcinomas, 3 cases showed an ACT to GCT (Thr to Ala) transition at codon 58, a CGG to CAG (Arg to Gln) transition at codon 241 and a CCC to TCC (Pro to Ser) transition at codon 308. All four other cases showed TTC to TCC (Phe to Ser) transitions at codon 295. These results suggest that mutations of the Lpar1 gene may be involved in the acquisition of growth advantage from adenomas to adenocarcinomas during rat lung carcinogenesis induced by BHP20. By contrast, no mutation of the Lpar2, Lpar3, Lpar4 and Lpar5 genes was detected in this model21.
Liver: To evaluate the Lpar1 gene mutation during rat liver carcinogenesis, hepatocellular carcinomas (HCCs) were induced by exogenous and endogenous liver carcinogenesis models22–24. For the exogenous model, HCCs were induced by N-nitrosodiethylamine (DEN), which is one of the most well-known liver carcinogens. On the other hand, endogenous carcinogenesis means that tumors are induced by endogenous changes that occur without any established carcinogen exposure, and prolonged feedings of a choline-deficient L-amino acid-defined (CDAA) diet can induce rat HCCs24. Missense mutations of the Lpar1 gene were detected in 7 out of 15 HCCs (46.7%) induced by DEN . By contrast, no mutation of Lpar5 was found in HCCs induced by DEN. Five out of 12 HCCs (41.7%) induced by the CDAA diet showed missense mutations25. Therefore, it is suggested that the Lpar1 mutations may play an important role in the development of rat HCCs induced by both liver carcinogenesis models.
Pancreas: A number of human pancreatic cancers commonly arise from pancreatic duct cells. Hamster pancreatic duct adenocarcinomas (PDAs) induced by the rapid production model using N-nitrosobis(2-oxopropyl)amine (BOP) histologically and genetically resemble the human situation26,27. In hamster PDAs induced by this model, only one out of 10 cases (10%) showed a GGA to GTA (Gly to Val) transversion at codon 355 of the Lpar1 gene28. Therefore, it seems that the Lpar1 gene mutation may be involved in a limited fraction of BOP-induced pancreatic duct carcinogenesis in hamsters.
Cancer cell lines: No mutation of the Lpar1 gene was found in cell lines established from hamster PDAs induced by BOP28. In RLCNR rat lung adenocarcinoma cells, COS rat osteosarcoma cells, RH7777 rat hepatoma cells, B103 rat neuroblastoma and C6 glioma cells, no mutation of the Lpar5 was detected29(Table 1).
Table 1. Mutations of LPA Receptor Genes in Rodent Tumors.
Location of the Lpar1 gene mutations in rodent tumors
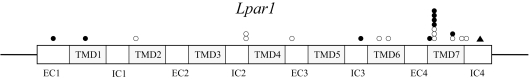
The Lpar1 gene mutations were observed in several positions in rodent tumors. Particularly, the frequency of missense mutations of codon 295 was high. In lung adenocarcinomas induced by BHP, 4 out of 7 mutations were located at codon 29520. In HCCs induced by exogenous and endogenous carcinogenesis, 3 out of 12 mutations were also found in codon 29525. The biological roles of mutations in codon 295 are unclear. However, codon 295 is adjacent to Lys294, which is one of the critical residues for ligand recognition or binding in the putative 7th transmembrane domain of LPA130. Recently, it has been reported that artificial replacement of Lys294 with Ala resulted in an enhancement of LPA response31. LPA2 and LPA3 contained basic amino acids equivalent to Lys294 of LPA1. These amino acids are thought to play an important role in LPA binding and the subsequent receptor activation. Therefore, these findings suggest that the mutations in codon 295 of the Lpar1 gene might affect the physical properties of the ligand recognition region in the 7th transmembrane domain, leading to changes in affinity of LPA binding or activation of signaling pathways upon LPA stimulation. The locations of Lpar1 gene mutations in lung, liver and pancreatic tumors of rodents are shown in Fig. 2.
Fig. 2.
The locations of Lpar1 gene mutations in lung, liver
and pancreatic tumors of rodents. EC, extracellular domain. TMD,
transmembrane domain. IC, intracellular domain.  , rat lung
adenomas and adenocarcinomas;
, rat lung
adenomas and adenocarcinomas; , rat
hepatocellular carcinomas;
, rat
hepatocellular carcinomas; , hamster
pancreatic duct adenocarcinomas.
, hamster
pancreatic duct adenocarcinomas.
Patterns of the Lpar1 gene mutations in rodent tumors
In tumors induced by nitrosocompounds, the Lpar1 gene mutation patterns have varied. Among 9 mutations in rat lung carcinogenesis model using BHP, 7 cases were T/A to C/G transitions and two were G/C to A/T transitions20. In DEN-induced rat HCCs, three T/A to C/G transitions, a C/G to T/A transition and three G/C to T/A transversions were observed25. In hamster PDAs induced by BOP, one mutation was a G/C to T/A transversion28. Although G/C to A/T transition is a common mutation pattern induced by nitrosocompounds32,33, only two lung adenocarcinomas and one HCC showed this pattern. By contrast, all 5 mutations in rat HCCs by the CDAA diet were T/A to C/G transitions25. Oxy radical-mediated DNA damage generates several DNA adducts, including 8-hydroxyguanine and 8-hydroxyadenine34. It is well established that 8-hydroxyguanine induces G/C to T/A or A/T to C/G transversions in Escherichia coli35. Moreover, 8-hydroxyadenine also induces a T/A to C/G transition or an A/T to C/G transversion in mammalian cells36,37. Since T/A to C/G transition is a frequent pattern, it is suggested that the Lpar1 gene may be a target for oxidative DNA damage in rodent tumors. In fact, it has been reported that oxidative stress is involved in rat hepatocarcinogenesis induced by the CDAA diet38.
Epigenetic Alterations
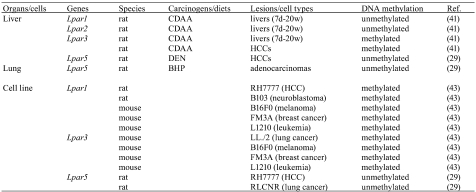
As one of epigenetic mechanisms for the regulation of gene expression, DNA methylation of cytosine residues at CpG dinucleotides is a strong mediator for the gene silencing in mammalian genomes39,40. It has been reported that loss of tumor suppressor gene expressions is due to aberrant DNA methylation of gene promoter regions in several tumors39,40. In LPA receptor genes, distinct expression and DNA methylation patterns of LPA receptor genes were found in human colon cancer cells16. Aberrant DNA methylation statuses of LPA receptor genes were also detected in rodent tumors (Table 2).
Table 2. Aberrant DNA Methylation of LPA Receptor Genes in Rodent Tumors and Cancer Cell Lines.
Liver
DNA methylation status and expression levels of the Lpar1, Lpar2 and Lpar3 genes were examined in a rat liver carcinogenesis model induced by the CDAA diet. The Lpar1 and Lpar2 genes were unmethylated in the livers of rats fed the CDAA diet for 7 days and 2, 12 and 20 weeks, as well as in normal liver tissues. In regard to the Lpar3 gene, the livers at 7 days and 2 and 12 weeks were weakly or moderately methylated, and those at 20 weeks were markedly methylated, while normal liver tissues were unmethylated. Moreover, 4 HCCs induced by the CDAA diet were completely methylated. In those samples, the expression levels of the Lpar1, Lpa2 and Lpar3 genes were correlated with the DNA methylation status41.
In regard to the Lpar5 gene, 4 out of 6 HCCs induced by DEN were unmethylated, compared with normal liver tissues, which were methylated. These DNA methylation patterns were correlated with the Lpar5 gene expression levels29.
Lung
In rat normal lung tissues, the Lpar5 gene was not expressed, and its DNA methylation status was methylated. By contrast, 5 out of 6 adenocarcinomas (83.3%) induced by BHP indicated increased expressions of Lpar5 with an unmethylated status29.
Cancer cell lines
While normal liver and brain tissues expressed the Lpar1 gene and its DNA methylation status was unmethylated, RH7777 and B103 cells showed hypermethylation of the Lpar1 gene with elevated expression levels42. In regard to the Lpar5 gene, RLCNR and RH7777 cells were unmethylated compared with normal lung and liver tissues29. In mouse tumor cells, aberrant DNA methylation of the Lpar1 gene was detected in B16F0 melanoma cells, FM3A mammary carcinoma cells and L1210 leukemia cells. The Lpar3 gene in LL/2 lung tumor cells, B16F0, FM3A and L1210 cells was also hypermethylated compared with their adjacent normal tissues43.
Conclusion
We here reviewed the genetic and epigenetic alterations of LPA receptor genes occurring in rodent tumors induced by experimental carcinogenesis models and cancer cell lines, demonstrating that alterations of the LPA signaling pathway may be involved in the development of tumor cells in rodents. Our successive study demonstrated that an artificial mutated form of LPA1 that lacked the carboxyl terminal was constitutively active and oncogenic44. Therefore, it is suggested that LPA receptors may be one of the important molecules for the development of anticancer and chemoprevention agents in clinical cancer approaches.
Acknowledgments
This study was supported in part by a Grant-in-Aid (20591765) for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, by Grants (21321201) from the Ministry of Health, Labor and Welfare of Japan and by grants from the Faculty of Science and Engineering, Kinki University.
References
- 1.Contos JJA, Ishii I, Chun J. Lysophosphatidic acid receptors. Mol Pharmacol. 58: 1188–1196 2000. [DOI] [PubMed] [Google Scholar]
- 2.Aoki J, Inoue A, Okudaira S. Two pathways for lysophosphatidic acid production. Biochim Biophys Acta. 1781: 513–518 2008. [DOI] [PubMed] [Google Scholar]
- 3.Ishii I, Fukushima N, Ye X, Chun J. Lysophospholipid receptors: Signaling and biology. Annu Rev Biochem. 73: 321–354 2004. [DOI] [PubMed] [Google Scholar]
- 4.Lin M-E, Herr DR, Chun J. Lysophosphatidic acid (LPA) receptors: Signaling properties and disease relevance. Prostaglandins Other Lipid Mediat. 91: 130–138 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang X, Schummer M, Mao M, Yu S, Tabassam FH, Swaby R, Hasegawa Y, Tanyi JL, LaPushin R, Eder A, Jaffe R, Erickson J, Mills GB. Lysophosphatidic acid is a bioactive mediator in ovarian cancer. Biochim Biophys Acta. 1582: 257–264 2002. [DOI] [PubMed] [Google Scholar]
- 6.Furui T, LaPushin R, Mao M, Khan H, Watt SR, Watt MAV. Overexpression of Edg-2/vzg-1 Induces apoptosis and anoikis in ovarian cancer cells in a lysophatidic acid-independent manner. Clinical Cancer Res. 5: 4308–4318 1999 [PubMed] [Google Scholar]
- 7.Goetzl EJ, Dolezalova H, Kong Y, Hu YL, Jaffe RB, Kalli KR, Conover CA. Distinctive expression and functions of the type 4 endothelial differentiation gene-encoded G protein-coupled receptor for lysophosphatidic acid in ovarian cancer. Cancer Res. 59: 5370–5375 1999. [PubMed] [Google Scholar]
- 8.Shida D, Watanabe T, Aoki J, Hama K, Kitayama J, Sonoda H, Kishi Y, Yamaguchi H, Sasaki S, Sako A, Konishi T, Arai H, Nagawa H. Aberrant expression of lysophosphatidic acid (LPA) receptors in human colorectal cancer. Lab Invest. 84: 1352–1362 2004. [DOI] [PubMed] [Google Scholar]
- 9.Schulte KM, Beyer A, Kohrer K, Oberhauser S, Roher HD. Lysophosphatidic acid, a novel lipid growth factor for human thyroid cells: over-expression of the high-affinity receptor edg4 in differentiated thyroid cancer. Int J Cancer. 92: 249–256 2001. [DOI] [PubMed] [Google Scholar]
- 10.Yu S, Murph MM, Lu Y, Liu S, Hall HS, Liu J, Stephens C, Fang X, and Mills GB. Lysophosphatidic acid receptors determine tumorigenicity and aggressiveness of ovarian cancer cells. J Natl Cancer Inst. 100: 1630–1642 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tabata K, Baba K, Shiraishi A, Ito M, and Fujita N. The orphan GPCR GPR87 was deorphanized and shown to be a lysophosphatidic acid receptor. Biochem Biophys Res Commun. 363: 861–866 2007. [DOI] [PubMed] [Google Scholar]
- 12.Ishii I, Contos JJA, Fukushima N, Chun J. Functional comparisons of the lysophosphatidic acid receptors, LPA1/VZG-1/EDG-2, LPA2/EDG-4, and LPA3/EDG-7 in neuronal cell lines using a retrovirus expression system. Mol Pharmacol. 58: 895–902 2000. [DOI] [PubMed] [Google Scholar]
- 13.Contos JJA, Ishii I, Fukushima N, Kingsbury MA, Ye X, Kawamura S, Brown JH, Chun J. Characterization of lpa(2) (Edg4) and lpa(1)/lpa(2) (Edg2/Edg4) lysophosphatidic acid receptor knockout mice: signaling deficits without obvious phenotypic abnormality attributable to lpa(2). Mol Cell Biol. 22: 6921–6929 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimomura Y, Wajid M, Ishii Y, Shapiro L, Petukhowa L, Gordon D, Christiano AM. Disruption of P2RY5, an orphan G protein-coupled receptor, underlies autosomal recessive woolly hair. Nature Genet. 40: 335–339 2008. [DOI] [PubMed] [Google Scholar]
- 15.Pasternack SM, von Kügelgen I, Aboud KAJ, Lee Y-A, Rüschendorf F, Voss K, Hillmer AM, Molderings GJ, Franz T, Ramirez A, Nürnberg P, Nöthen MM, Berz RC. G protein-coupled receptor P2Y5 and its ligand LPA are involved in maintenance of human hair growth. Nature Genet. 40: 329–334 2008. [DOI] [PubMed] [Google Scholar]
- 16.Tsujino M, Fujii M, Okabe K, Mori T, Fukushima N, Tsujiuchi T. Differential expressions and DNA methylation patterns of lysophosphatidic acid receptor genes in human colon cancer cells. Virchows Arch. 457: 669–676 2010. [DOI] [PubMed] [Google Scholar]
- 17.Okabe K, Hayashi M, Fujii M, Honoki K, Mori T, Fukushima N, Tsujiuchi T. Mutations of lysophosphatidic acid receptor genes in human osteosarcoma cells. Pathobiology. 77: 278–282 2010. [DOI] [PubMed] [Google Scholar]
- 18.Konishi Y, Denda A, Kondo H, Takahashi S. Lung carcinomas induced by oral administration of N-bis(2-hydroxypropyl)nitrosamine in rats. Gann. 67: 773–780 1976. [PubMed] [Google Scholar]
- 19.Konishi Y, Kondoh H, Denda A, Takahashi S, Inui S. Lung carcinomas induced by oral administration of N-bis(2- hydroxypropyl)nitrosamine in rats. In: Tumors of Early Llife in Man and Animals. Perugia Quadrennaial International Conference of Cancer. I Severi (ed.) Division of Cancer Resarch Perugia University, Perugia. 637–649. 1978
- 20.Yamada T, Furukawa M, Hotta M, Yamasaki A, Honoki K, Fukushima N, Tsujiuchi T. Mutations of lysophosphatidic acid receptor-1 gene during progression of lung tumors in rats. Biochem Biophys Res Commun. 378: 424–427 2009. [DOI] [PubMed] [Google Scholar]
- 21.Wakabayashi N, Tsujino M, Tajiri M, Taki M, Koshino A, Ikeda H, Fukushima N, Tsujiuchi T. No mutations of lysophosphatidic acid receptor genes in lung adenocarcinomas induced by N-nitrosobis(2-hydroxypropyl)amine in rats. J Toxicol Pathol. 23: 63–66 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohashi K, Tsutsumi M, Tsujiuchi T, Kobitsu K, Okajima E, Nakajima Y, Nakano H, Takahashi M, Mori Y, Konishi Y. Enhancement of N- nitrosodiethylamine-initiated hepatocarcinogenesis caused by a colchicines-induced cell cycle disturbance in partially hepatectomized rats. Cancer Res. 56: 3474–3479 1996 [PubMed] [Google Scholar]
- 23.Tsutsumi M, Ohashi K, Tsujiuchi T, Kobayashi E, Kobitsu K, Kitada H, Majima T, Okajima E, Endoh T, Hasegawa K, Mori T, Konishi Y. Disturbance of the cell cycle with colchicines enhances the growth advantage of diethylnitrosamine-induced hepatocytes in rats. Jpn J Cancer Res. 87: 5–9 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakae D, Yoshiji H, Mizumoto Y, Horiguchi K, Shiraiwa K, Tamura K, Denda A, Konishi Y. High incidence of hepatocellular carcinomas induced by a choline deficient L-amino acid defined diet in rats. Cancer Res. 52: 5042–5045 1992. [PubMed] [Google Scholar]
- 25.Obo Y, Yamada T, Furukawa M, Hotta M, Honoki K, Fukishima N, Tsujiuchi T. Frequent mutations of lysophosphatidic acid receptor-1 gene in rat liver tumors. Mutat Res. 660: 47–50 2009. [DOI] [PubMed] [Google Scholar]
- 26.Mizumoto K, Tsutsumi M, Denda A, Konishi Y.Rapid production of pancreatic carcinoma by initiation with N-nitrosobis(2-oxopropyl)amine and repeated augmentation pressure in hamsters. J Natl Cancer Inst. 80: 1564–1567 1988. [DOI] [PubMed] [Google Scholar]
- 27.Mizumoto K, Kitazawa S, Ito S, Takashima Y, Tsutsumi M, Denda A, Konishi Y.Cycles of repeated augmentation pressure in rapid production of pancreatic and cholangiocellular carcinomas in hamsters initiated with N-nitrosobis(2-oxopropyl)amine. Carcinogenesis 10: 1457– 1459 1989. [DOI] [PubMed] [Google Scholar]
- 28.Tsujiuchi T, Furukawa M, Yamasaki A, Hotta M, Kusunoki C, suyama N, Mori T, Honoki K, and Fukushima N. Infrequent mutation of lysophosphatidic acid receptor-1 gene in hamster pancreatic duct adenocarcinomas and established cell lines. J Toxicol Pathol. 22: 89–92 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okabe K, Hayashi M, Yamawaki Y, Teranishi M, Honoki K, Mori T, Fukushima N, Tsujiuchi T. Possible involvement of lysophosphatidic acid receptor-5 gene in the acquisition of growth advantage of rat tumor cells. Mol Carcinog. 2011; . [DOI] [PubMed] [Google Scholar]
- 30.Sardar VM, Bautista DL, Fischer DJ, Yokoyama K, Nusser N, Viraq T, Wang DA, Baker DL, Tiqyi G, Parrill AL. Molecular basis for lysophosphatidic acid receptor antagonist selectivity. Biochim Biophys Acta. 1582: 309–317 2002. [DOI] [PubMed] [Google Scholar]
- 31.Valentine WJ, Fells JI, Perygin DH, Mujahid S, Yokoyama K, Fujiwara Y, Tsukahara R, Van Brocklyn JR, Parrill AL, Tiqyi G. Subtype-specific residues involved in ligand activation of the endothelial differentiation gene family lysophosphatidic acid receptors. J Biol Chem. 283: 12175–12187 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitada H, Tsutsumi M, Tsujiuchi T, Takahama M, Fukuda T, Narita N, Konishi Y. Frequent mutations of Ki-ras but no mutations oh Ha-ras and p53 in lung lesions induced by N-nitrosobis(2-hydroxypropyl)amine in rats. Mol Carcinog. 15: 276–283 1996. [DOI] [PubMed] [Google Scholar]
- 33.Tsutsumi M, Murakami Y, Kondoh S, Tsujiuchi T, Honoki K, Horiguchi K, Noguchi O, Kobayashi E, Okita S, Sekiya T, Konishi Y. Comparison of K-ras oncogene activation in pancreatic duct carcinomas and cholangiocarcinomas induced in hamsters by N-nitrosobis(2-oxopropyl)amine. Jpn J Cancer Res. 84: 956–960 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malins DC, Haimanot R. 4,6-Diamino-5-formamidopyrimidine, 8-hydroxyguanine and 8-hydroxyadenine in DNA from neoplastic liver of English sole exposed to carcinogens. Biochem Biophys Res Commun. 173: 614–619 1990. [DOI] [PubMed] [Google Scholar]
- 35.Moriya M, Ou C, Bodepudi V, Johnson F, Takeshita M, Grollma AP. Site-specific mutagenesis using a gapped duplex vector: a study of translation synthesis past 8-oxodeoxyguanosine in E. coli. Mutat Res. 254: 281–288 1991. [DOI] [PubMed] [Google Scholar]
- 36.Kamiya H, Miura H, Murata-Kamiya N, Ishikawa H, Sakaguchi T, Inoue H, Sasaki T, Masutani C, Hanaoka F, Nishimura S. 8-Hydroxyadenine (7,8-dihydro-8-oxoadenine) induces misincorporation in in vitro DNA synthesis and mutations in NIH3T3 cells. Nucleic Acids Res. 23: 2893–2899 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D, Kreutzer DA, Essigmann JM. Mutagenicity and repair of oxidative DNA damage: insights from studies using defined lesions. Mutat Res. 400: 99–115 1998. [DOI] [PubMed] [Google Scholar]
- 38.Nakae D, Yoshiji H, Maruyama H, Kinugasa T, Denda A, Konishi Y. Production of both 8-hydroxydeoxyguanosine in liver DNA and γ-glutamyltransferase-positive hepatocellular lesions in rats given a choline-deficient, L-amino acid-defined diet. Jpn J Cancer Res. 81: 1081–1084 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones PA. DNA methylation and cancer. Oncogene. 21: 5358–5360 2002. [DOI] [PubMed] [Google Scholar]
- 40.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 3: 415–428 2002. [DOI] [PubMed] [Google Scholar]
- 41.Okabe K, Hayashi M, Yoshida I, Nishimura K, Fukushima N, Tsujiuchi T. Distinct DNA methylation patterns of lysophosphatidic acid receptor genes during rat hepatocarcinogenesis induced by a choline deficient L-amino acid defined diet. Arch Toxicol. 2011. [DOI] [PubMed] [Google Scholar]
- 42.Tsujiuchi T, Shimizu K, Onishi M, Sugata E, Fujii H, Mori T, Honoki K, Fukushima N. Involvement of aberrant DNA methylation on reduced expression of lysophosphatidic acid receptor-1 gene in rat tumor cell lines. Biochem Biophys Res Commun. 349: 1151–1155 2006. [DOI] [PubMed] [Google Scholar]
- 43.Okabe K, Hayashi M, Wakabayashi N, Yamawaki Y, Teranishi M, Fukushima N, Tsujiuchi T. Different expressions and DNA methylation patterns of lysophosphatidic acid receptor genes in mouse tumor cells. Pathobiology. 77: 309–314 2010. [DOI] [PubMed] [Google Scholar]
- 44.Shano S, Hatanaka K, Ninose S, Moriyama R, Tsujiuchi T, Fukushima N. A Lysophosphatidic acid receptor lacking the PDZ-binding domain is constitutively active and stimulates cell proliferation. Biochim Biophys Acta. 1783: 748–759 2008. [DOI] [PubMed] [Google Scholar]