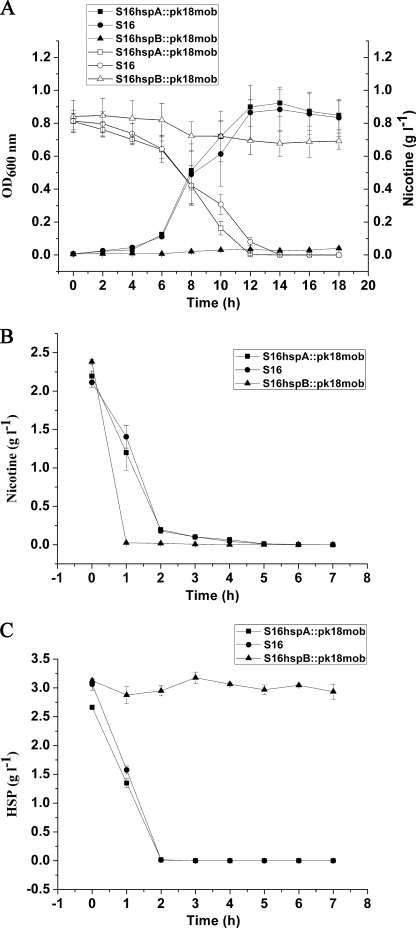
FIGURE 7.
Cell growth and resting cell reactions of strain S16 and the gene deletion mutants. A, utilization of nicotine for growth by S16 hspA::pk18mob (deletion of hspA), S16 (as control), and S16 hspB::pk18mob (deletion of hspB). ■, cell growth of S16 hspA::pk18mob (deletion of hspA); ●, cell growth of S16 as control; ▲, cell growth of S16 hspB::pk18mob (deletion of hspB); □, nicotine concentration in the medium for S16 hspA::pk18mob growth; ○, nicotine concentration in the medium for S16 growth as control; △, nicotine concentration in the medium for S16 hspB::pk18mob growth. B, resting cell reactions of S16 hspA::pk18mob (deletion of hspA), S16 (as control), and S16 hspB::pk18mob (deletion of hspB) in nicotine degradation. ■, nicotine concentration in the medium for S16 hspA::pk18mob degradation; ●, nicotine concentration in the medium for S16 degradation as control; ▲, nicotine concentration in the medium for S16 hspB::pk18mob degradation. C, resting cell reactions of S16 hspA::pk18mob (deletion of hspA), S16 (as control), and S16 hspB::pk18mob (deletion of hspB) in HSP degradation. ■, HSP concentration in the medium for S16 hspA::pk18mob degradation; ●, HSP concentration in the medium for S16 degradation as control; ▲, HSP concentration in the medium for S16 hspB::pk18mob degradation. The values are the means of three replicates, and the error bars indicate the standard deviations.