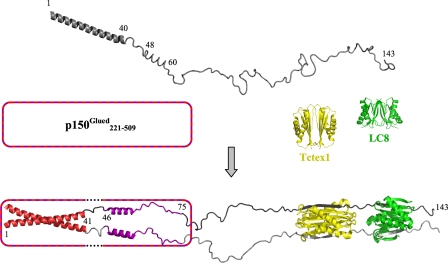
FIGURE 8.
Model for assembly of dynein intermediate chain with p150Glued and light chains. The model depicts apo-IC:1–143 (top) as a primarily disordered and monomeric ensemble of conformations with one defined helical region encompassing the N-terminal 40 residues and another region of nascent helicity encompassing residues ∼48–60, as determined from NMR measurements. Segments of IC for which NMR spectral characteristics are altered by p150Glued binding are indicated in red (region 1, residues 1–41) and purple (region 2, residues 46–75). The helical nature of regions 1 and 2 in bound IC (bottom) is inferred as described in the text, whereas all other regions in the bound complex are directly measured by NMR spectroscopy or x-ray crystallography. In accordance with sequence-based prediction of coiled-coil conformations in both the N-terminal α-helical region of IC and the p150Glued construct used in this study, the N-terminal 40 residues of IC in the bound state are depicted in a generic coiled-coil configuration. The p150Glued·IC complex is a tetramer that could be formed either from two p150Glued/IC heterodimeric coiled-coils (i.e. one chain from IC and one chain of p150Glued) or from two homodimeric coiled-coils (i.e. two chains of IC/IC coiled-coil packed against two chains of p150Glued coiled-coil). The p150Glued construct is represented as a box with a striped border. In the IC-bound state, dotted lines in the box indicate that p150Glued does not interact with IC residues 42–45 (linker 1). Residues in linker 1 and linker 2 (residues 76–106) are disordered, as indicated by NMR (Fig. 7). The IC·Tctex1·LC8 subcomplex portion of the model is from a crystal structure (Protein Data Bank entry 3FM7) (24), with Tctex1 (yellow) and LC8 dimers (green), and the corresponding IC segments in gray (one subunit light gray and the other dark gray). All structures, including apo-Tctex1 (Protein Data Bank entry 1YGT) and apo-LC8 (Protein Data Bank entry 3BRI) were generated using PyMOL (51).