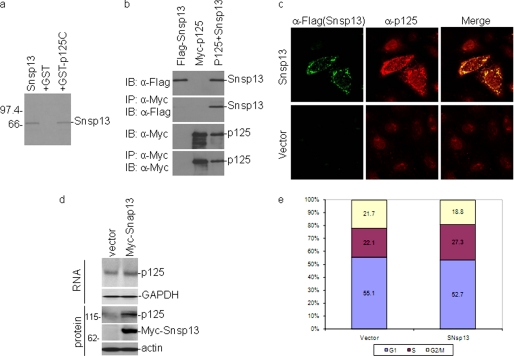
FIGURE 8.
Interaction of SARS-CoV nsp13 with the p125 subunit of Pol δ and induction of DNA damage response and cell cycle arrest. a, physical association of SARS-CoV nsp13 (Snsp13) with p125C in GST pulldown assay is shown. GST and GST-p125C were used to pull down the 35S-labeled, in vitro translated Snsp13. The precipitates and the in vitro translation products were detected by autoradiography. The GST protein was used as a negative control. b, shown is co-immunoprecipitation (IP) of Snsp13 with full-length p125 in mammalian cells overexpressing the two proteins. HeLa cells were transiently transfected with DNA constructs coding for FLAG-Snsp13 (first lane 1), myc-p125 (second lane), or both (third lane). Cells were lysed with lysis buffer at 28 h post-transfection, and co-immunoprecipitation was carried out using specific antibodies against Myc. The precipitates and total cell lysates were immunoblotted (IB) with indicated antibodies. c, partial relocalization of p125 from the nucleus to the cytoplasm in cells overexpressing Snsp13 is shown. HeLa cells were transfected with plasmid pXJFLAG-Snsp13 or empty vector pXJFLAG and fixed at 24 h post-transfection. Cells were co-immunostained with mouse anti-p125 and rabbit anti-FLAG antibodies. Red and green represent endogenous p125 and the FLAG-tagged Snsp13, respectively. Cells transfected with an empty vector expressing the FLAG tag alone were used as a negative control. d, shown is a Western blot analysis of cells stably expressing Snsp13. The expression of the endogenous p125 and Myc-Snsp13 in Snsp13-expressing stable cells was determined by Western blot analysis with anti-p125 and anti-Myc antibodies. The expression of the endogenous p125 and GAPDH at the mRNA level in Snsp13-expressing cells was determined by Northern blot analysis. e, induction of S-phase arrest in cells stably expressing SARS-CoV nsp13 is shown. Cell cycle profiles of stable Snsp13-expressing cell clones (SNsp13) and control cells (vector) were determined by flow cytometry analysis with PI staining. Unsynchronized cells grown for 24 h were fixed and stained for FACS analysis. A total of 10,000 cells were counted in each experiment, and the results represent the means of seven repeated experiments. Statistical analysis was carried by t test (p < 0.001, n = 7).