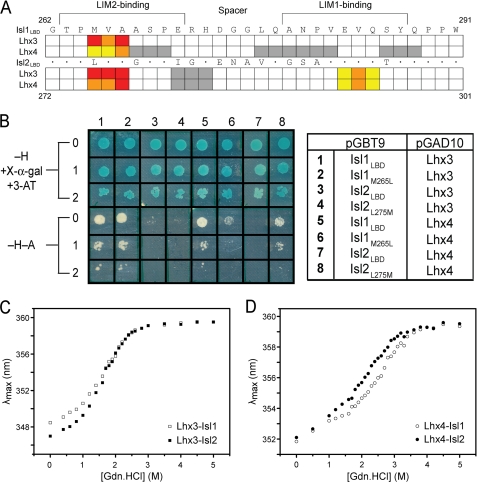
FIGURE 6.
Binding and apparent stability in Lhx3/4·Isl1/2 complexes. A, summary of alanine mutagenic screening assayed by yeast two-hybrid analysis from supplemental Table S4. The sequence of Isl1LBD (262–291) is shown. The sequence of Isl2LBD (272–301) shows where residues are conserved (*) or different. Colored boxes indicate where mutation had a strong (red), moderate (orange), or minor effect (yellow). White boxes indicate residues mutated in triple-alanine constructs that had a minor effect on growth of yeast. Gray boxes indicate no effect on yeast growth. B, comparison of wild-type and mutant LBDs from Isl1/Isl2 binding to the LIM domains of Lhx3/Lhx4 by yeast two-hybrid analysis. AH109 yeast cells co-transformed with pGBT9/pGAD10 vectors shown on the right were tested for growth under different selection conditions (−H + X-α-gal + 3-AT) or (−H−A); 0 indicates no dilution of yeast cells (A600 nm = 0.2), 1 indicates a 1:10 dilution (A600 nm = 0.02), and 2 indicates a 1:100 dilution (A600 nm = 0.002). C, resistance of Lhx3-Isl1/2 complexes to denaturation by guanidine hydrochloride (Gdn.HCl). □, Lhx3-Isl1; ■, Lhx3-Isl2. D, resistance of Lhx4-Isl1/2 complexes to denaturation by guanidine hydrochloride. ○, Lhx4-Isl1; ●, Lhx4-Isl2. For C and D, λmax reports maximum emission wavelength in the range 320–380 nm with excitation at 295 nm.