FIGURE 2.
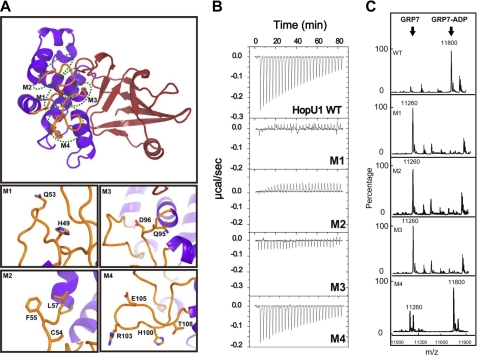
Effect of HopU1 mutations on activity and GRP7 recognition. A, structure of HopU1 indicating the location of HopU1 mutations M1–M4 (detailed in bottom panels). The residues changed to Ala are highlighted and shown as stick representations. The structure was rotated ∼90° about the horizontal axis compared with the view presented on the left side of Fig. 1A. B, isothermal titration calorimetry of wild-type HopU1 and HopU1 derivatives M1–M4 with GRP7, indicating the extent that the HopU1 derivatives can interact with GRP7. C, electrospray ionization-mass spectrometry results of mADP-RT activity for wild-type HopU1 and HopU1 derivatives. The peak of substrates and products of each enzymatic reaction are indicated. The molecular mass of GRP7 (1–90 with 4 additional residues (Gly, Pro, His, and Met) left over after affinity tag cleavage) is 11,260 Da and GRP7 with an ADP-ribose modification is 11,800 Da. The mutations of each HopU1 mutant are as follows: M1, HopU1H49A/Q53A; M2, HopU1C54A/F55A/L57A; M3, HopU1Q95A/D96A; and M4, HopU1H100A/R103A/E105A/T108A.