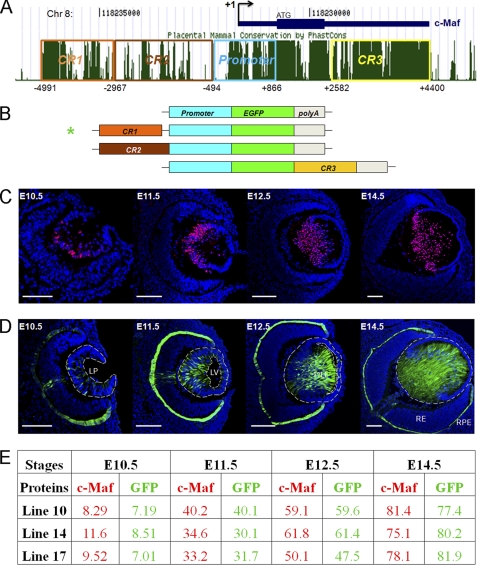
FIGURE 1.
Identification and functional characterization of a novel 5′ lens-preferred enhancer in the c-Maf locus. A, evolutionarily conserved regions (CR1, 2, and 3) in the 12-kb region of the mouse c-Maf locus. Mammal Cons displays the conservation of DNA in multiple mammalian species shown in the University of California at Santa Cruz genome browser. B, a diagrammatic summary of four EGFP reporter constructs tested in transgenic mice. The individual conserved regions CR1 (1.6 kb), CR2 (2.5 kb), and CR3 (1.8 kb) were tested in combination with a 1.3 kb c-Maf promoter (-500 to +800). C, expression of c-Maf in the mouse embryonic lens detected by immunofluorescence. D, expression of EGFP driven by CR1/1.3 kb c-Maf promoter in the transgenic mouse eye (line 10). Expression of c-Maf in RPE by in situ hybridizations and using the knocked-in lacZ marker were described elsewhere (23, 24). LP, lens pit; LV, lens vesicle; 1oLF, primary lens fibers; RE, retina; RPE, retinal pigmented epithelium. Scale bars = 100 μm (C and D). Blue, DAPI; red, anti-c-Maf; green, anti-GFP. E, quantitative analysis of c-Maf- and EGFP-expressing cells in three independent transgenic lines (lines 10, 14, and 17; see C and D and supplemental Fig. S1). The percentages of c-Maf- and EGFP-positive cells were calculated as described under “Experimental Procedures.”