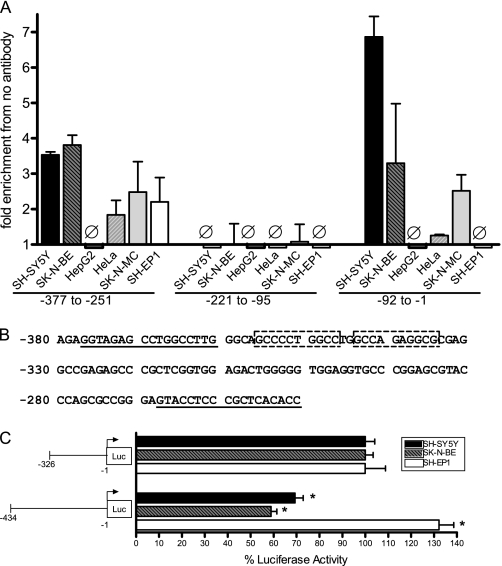
FIGURE 6.
Chromatin immunoprecipitation identification of AP-2α binding sites in the CHRNA7 promoter. A, SH-SY5Y, SK-N-BE, SK-N-MC, HeLa, HepG2, or SH-EP1 cells were formaldehyde-cross-linked and lysed, and lysates were sonicated to obtain DNA fragments 200–300 bp in length. Immunoprecipitation was performed using Sepharose beads and anti-AP-2α antibody or no antibody plus lysate. DNA fragments isolated by immunoprecipitation were used for quantitative PCR to amplify relevant CHRNA7 promoter sequences as indicated, using the primers shown in Table 1. Fold enrichment compared with no antibody control is indicated. Ø, no change from controls. Triplicates were used for each experiment, and experiments were replicated in triplicate. Data are presented as mean ± S.E. (error bars). B, a second region containing AP-2α binding was identified using chromatin immunoprecipitation. Two putative AP-2α consensus binding sites are indicated by boxes within the CHRNA7 promoter region. The positions of primers ChIP-315F and ChIP-315R are underlined. C, transient transfection of SH-SY5Y, SK-N-BE, and SH-EP1 cells with luciferase reporter vectors containing 5′ deletions of the CHRNA7 promoter at the second putative AP-2α binding region, normalized to Renilla expression from the pRL-TK vector. Eight wells were used for each experiment, and experiments were replicated in triplicate. −326 was set to 100% for each cell line. Data are presented as mean ± S.E. *, p < 0.0005.