Background: SmTK6 was identified as interaction partner of SmTK4.
Results: SmTK6 is a Src/Abl hybrid kinase and interacts also with the uncommon SmVKR1 and SmTK3.
Conclusion: SmTK6 is suggested to be part of a complex of receptors, Syk and Src kinases, which are involved in gonad development.
Significance: SmTK6 represents an Abl kinase progenitor, for which a function in reproduction could be assigned.
Keywords: Cell Differentiation, Oocyte, Parasite, Signal Transduction, Src, Abl, Schistosoma, Syk, Venus Kinase Receptor, Kinase Inhibitor
Abstract
Cellular protein-tyrosine kinases play key roles in signal transduction processes in eukaryotes. SmTK4 was the first Syk kinase identified in a parasite and found to be tissue-specifically transcribed in the gonads of adult Schistosoma mansoni. Functional analyses confirmed its role in oogenesis and spermatogenesis. As an SmTK4 upstream binding partner, the cellular protein-tyrosine kinase SmTK6 was isolated from a yeast two-hybrid library. Phylogenetic analyses performed in this study confirmed the first suggestions of a hybrid character of SmTK6. Biochemical studies made in Xenopus oocytes using inhibitors against Src (herbimycin A) and Abl (imatinib) kinases exhibited a biochemical inhibition profile of SmTK6, which was intermediate of Src and Abl kinases. As SmTK6 upstream interaction partners, we identified among others the known Src kinase SmTK3 and the Venus kinase receptor SmVKR1 of S. mansoni by yeast two-hybrid analyses, all of which co-localized in the gonads. Co-immunoprecipitation experiments confirmed interactions between SmTK6 and SmTK3 or SmVKR1. In Xenopus oocytes, it was finally shown that SmVKR1 but also SmTK3 were able to activate SmTK6 enzymatic activity indicating its functions in a receptor tyrosine kinase signal transduction cascade. These results not only demonstrate an intermediate but Src-biased profile of the unusual kinase SmTK6. They also strongly substantiate previous indications for a kinase complex, consisting of a receptor tyrosine kinase, Syk and Src kinases, which has been hypothesized to be involved in proliferation and differentiation processes in the gonads of schistosomes.
Introduction
As essential members of signal transduction cascades, cellular protein-tyrosine kinases (CTKs)2 are involved in processes regulating cytoskeletal reorganization, migration, proliferation, development and differentiation, metabolic homeostasis, transcriptional activation, neural transmission, aging, and survival (1). CTKs forward incoming signals from transmembrane receptors to binding partners acting downstream in a signaling hierarchy (2, 3). Depending on their function and state of activation, CTKs can be either located in the cytoplasm, be attached to membranes, or occur in the nucleus (4). According to their structural features, CTKs were classified as the following distinct families: Src, Abl, Syk, Jak, Fak, Fes/Fer, Csk, and Btk (5). Among these, Src and Abl kinases have Src homology 3 and 2 (SH3 and SH2) domains and a catalytic tyrosine kinase (TK) domain in the same linear order. Src kinases possess two conserved regulatory tyrosine (Tyr) phosphorylation sites (1, 5). Phosphorylation of Tyr-527 within the C terminus is important for the inactive conformation of Src, mediated by intramolecular binding to its SH2 domain, whereas phosphorylation of Tyr-416, an autophosphorylation site within the activation loop of the TK domain, leads to the stimulation of kinase activity. SH2 and SH3 domains have been implicated in the negative regulation of Src activity, but both also contribute to the binding of partners acting upstream (via SH2) or downstream (via SH3) of Src in signaling cascades, if this kinase becomes activated (5). Regulation of Abl kinases differs as they lack Tyr-527 within the C terminus, which has no functional role in the control of Abl kinase activity. Instead, the inactive conformation is formed by intramolecular binding of a short N-terminal cap peptide, supported by a myristoyl group attached to the N-terminal glycine and to the TK domain. Downstream of the cap peptide are SH3 and SH2 domains contributing by interaction with the TK domain to the “locked” inactive state (6). Further features of Abl kinases from higher eukaryotes are the presence of a nuclear localization sequence and an elongated C-terminal region containing varying sets of binding domains for actins, microtubules, or DNA (6).
Schistosomes are helminth parasites causing schistosomiasis, one of the most prevalent parasitic diseases for human and animals worldwide (7, 8). Its pathology is directly associated with egg production of mature adult worms. Eggs cause granuloma formation and inflammatory processes, which interfere with organ function. The course of the disease can be fatal, affecting millions of people (9). As the only class within the trematodes, schistosomes have evolved separate sexes. Furthermore, by a continuous pairing contact with the male, proliferation and differentiation processes are initiated in female gonads (10–12). This is the prerequisite for the synthesis of composite eggs consisting of oocytes originating from the ovary and vitelline cells delivered from the vitellarium. During the last years, first molecules have been characterized that are involved in regulating mitogenic activity and differentiation in female gonads. Among these are the Src kinase SmTK3 and the Syk kinase SmTK4 (13, 14). SmTK3 contains all Src-typical features, and expression products were localized in vitelline cells, oocytes, and spermatocytes suggesting a role in reproductive activity (15). Studies with the Src kinase-specific inhibitor herbimycin A provided strong evidence for a role of SmTK3 in the control of mitotic activity and egg production of adult schistosomes in vitro (16). Characterized by its tandem SH2 domains, SmTK4 is a typical Syk kinase. SmTK4 transcripts were found in spermatocytes and oocytes but not in vitelline cells (17). Using the Syk kinase-specific inhibitor piceatannol and RNAi knockdown approaches in adult worms in vitro demonstrated a decisive role of SmTK4 in oogenesis and spermatogenesis (13). The SmTK4 upstream interaction partner SmTK6 was identified and co-localized in the reproductive organs. Co-immunoprecipitation experiments confirmed direct interactions between both kinases (13). First database analyses comparing SmTK6 with two recently detected Abl kinases from schistosomes suggested that SmTK6 may represent an Src-/Abl-like hybrid kinase (18).
In this study, we provide functional evidence for the intermediate Src/Abl kinase characteristic of SmTK6 by gene structure and phylogenetic analyses and also by inhibitor studies. Furthermore, we identified upstream-binding partners in S. mansoni such as SmTK3, SmVKR1, a Drosophila Discs-large homolog (DLG), and a new transmembrane mucin. Transcripts of all these genes co-localized in the reproductive organs. Following co-immunoprecipitation experiments, which confirmed SmTK6-SmTK3 as well as SmTK6-SmVKR1 interactions, germinal vesicle breakdown (GVBD) assays in Xenopus oocytes finally demonstrated that SmTK6 can be activated by SmVKR1 or SmTK3. These results reinforce previous suggestions of a multikinase complex in the gonads of schistosomes consisting of the Syk kinase SmTK4, the Src kinase SmTK3, and the RTK SmVKR1, in which the unusual Src/Abl-like kinase SmTK6 is a novel player.
EXPERIMENTAL PROCEDURES
Parasite Stock
Adult and larval schistosome stages originated from a Liberian isolate of Schistosoma mansoni (19), which was maintained in snails (Biomphalaria glabrata) and Syrian hamsters (Mesocricetus auratus). Adult worms were obtained by hepatoportal perfusion at 42–49 days post-infection. Experiments with hamsters were performed in accordance with the European regulations (ETS 123; revised Appendix A) and were approved by the Regional Council, Giessen, Germany.
Yeast Two-hybrid Screening
A Y2H cDNA-library based on RNA of mixed sex adult S. mansoni (20) was used for the identification of SmTK6 upstream interaction partners. In this library, the cDNAs were cloned into the prey vector pGADT7-Rec (leucine nutritional marker LEU2, Clontech) in-frame with the GAL4 activation domain (GAL4-AD). Two yeast strains were used for screening, the library-containing strain AH109 (Mat a; reporter genes ADE2, HIS3, and LacZ) and the bait-containing strain Y187 (Matα; reporter genes HIS3 and LacZ). For library screening, a bait plasmid (pBridge, tryptophan nutritional marker TRP1; Clontech) was cloned containing the SH2 domain of SmTK6 within the MCS I in-frame with the GAL4 DNA-binding domain (GAL4-BD). The encoding sequence was amplified by PCR using the primer pair SmTK6-SH2–5′ (5′-GGATCCGTCTGAATGATGGACTTCCAACTAGTTTG-3′; containing a BamHI site) and SmTK6-SH2–3′(5′-CTGCAGAAATGCACTGGTGGACGGTATGC-3′; containing a PstI site), and a full-length cDNA clone of SmTK6 as template. The expected amplification product (355 bp) was obtained and cloned via BamHI/PstI into pBridge. After cloning, the resulting construct SmTK6-SH2 pBridge was sequenced confirming the correct open reading frame (ORF) of the GAL4-BD/SmTK6-SH2 fusion.
Library screening was performed according to the user manual (Yeast Protocols Handbook from Clontech). In short, yeast cells (strain Y187) were transformed with the bait plasmid SmTK6-SH2 pBridge by lithium acetate. Bait-expressing Y187 cells were mated with the library containing AH109 cells. The first selection of diploid yeast cells was carried out on synthetic dropout medium lacking tryptophan, leucine, and histidine (Trp−/Leu−/His−). To enhance the selection pressure on clones with interacting proteins, colonies were plated onto synthetic dropout medium additionally lacking adenine (Trp−/Leu−/His−/Ade−). For further selection, β-galactosidase (β-gal) colony filter assays were performed using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as substrate according to the manufacturer's instructions (Clontech). From positively tested yeast clones, plasmid DNA was isolated using cell disruption by vortexing with glass beads (Sigma) followed by plasmid preparation (peqGOLD plasmid mini kit, PeqLab). Plasmid DNA was transformed into heat shock-competent Escherichia coli cells (DH5α) followed by selection on LB plates containing ampicillin (100 μg/μl). To differentiate bacterial colonies containing bait plasmids from those containing prey plasmids, colony PCRs with pGADT7-specific primers were performed. Prey plasmids from PCR-positive bacterial clones were isolated and sequenced commercially (LGC Genomics, Berlin, Germany). To confirm protein-protein interactions, the yeast strain AH109 was transformed with appropriate prey plasmids together with the bait plasmid, and the selection procedures were repeated. For quantification of relative interaction strengths, β-gal liquid assays with o-nitrophenol galactopyranoside as substrate were performed according to conventional protocols (Clontech).
Direct Yeast Two-hybrid Interactions Studies
AH109 yeast cells (Mat a; reporter genes ADE2, HIS3, and LacZ) were co-transformed with appropriate bait and prey plasmids by lithium acetate. As prey vector pACT2 (leucine nutritional marker LEU2, GAL4-AD; Clontech) was used, and pBridge as bait vector was used. The intracellular region of the SmVKR1 was amplified using the primers VKR1-pACT2–5′ (5′-GGATCCTTTACTATCGGCGCAAACTAAAG-3′; containing a BamHI site) + VKR1-pACT2–3′ (5′-CTCGAGAAGGTAGAAACGCTAAACTGTTATC-3′; containing a XhoI site), and a full-length cDNA clone of SmVKR1 as template. The amplicon (1794 bp) was cloned via BamHI/XhoI into pACT2. After cloning, the resulting construct SmVKR1-C-term pACT2 was sequenced confirming the correct ORF of the GAL4-AD/SmVKR1-C-term fusion. As bait plasmids, the following constructs were used, which express the relevant protein interaction domains of the schistosome CTKs SmTK6, SmTK3, and SmTK4 as a fusion with the GAL4-BD: SmTK6-SH3SH2 pBridge, SmTK6-SH3 pBridge, SmTK6-SH2 pBridge, SmTK3-SH3 SH2 pBridge, SmTK3-SH3 pBridge, SmTK3-SH2 pBridge, and SmTK4-SH2SH2 pBridge. Cloning of the bait construct SmTK4-SH2 SH2 pBridge was described elsewhere (13). For the amplification of the combined or individual SH3/SH2 domains of SmTK6 or SmTK3 by PCR, the following primer combinations were used: TK6-SH3SH2–5′ (5′-GGATCCGTATGGGAATTTGTTTGTGTCTTC-3′; containing a BamHI site) + TK6-SH3SH2–3′ (5′-CTGCAGAAGCTTTCGAATTCTTGTTATG-3′; containing a PstI site); TK6-SH3–5′ (5′-GGATCCGTTTGGTACAGGTTCGCGCTA-3′; containing a BamHI site) + TK6-SH3–3′ (5′-CTGCAGTCAAGTTGGAAGTCCATCATTCAG-3; containing a PstI site); TK3-SH3SH2–5′ (5′-GGATCCGTATGGGAAATTCTAATTCGTCTAA-3′; containing a BamHI site) + TK3-SH3SH2–3′ (5′-CTGCAGAATTGAAGATTTTGGAATTTCCC-3′; containing a PstI site); TK3-SH3–5′ (5′-GGATCCGTACAGAAGGGCAGTTTGTTGC-3′; containing a BamHI site) + TK3-SH3–3′ (5′-CTGCAGTCATTCCAAACTGGTAACAGCTG-3′; containing a PstI site), and TK3-SH2–5′ (5′-GGAGAATTCGAATGGTATTTTGGAG-3′; containing an EcoRI site) + TK3-SH2–3′ (5′-TCCGTCGACCGGTTTTCCCAATCGAC-3′; containing a SalI site). Amplification products of the expected sizes (TK6-SH3SH2, 923 bp; TK6-SH3, 206 bp; TK3-SH3SH2, 1052 bp; TK3-SH3, 206 bp; TK3-SH2, 309 bp) were cloned via BamHI/PstI or EcoRI/SalI in case of SmTK3-SH2 into pBridge. The resulting constructs SmTK6-SH3SH2 pBridge, SmTK6-SH3 pBridge, SmTK3-SH3SH2 pBridge, SmTK3-SH3 pBridge, and SmTK3-SH2 pBridge were sequenced confirming their correct ORFs.
Xenopus Oocyte Experiments and Inhibitor Studies
Sequences of TK domains of SmTK3, SmTK6, and SmAbl1 were obtained by PCR amplification using the respective full-length kinase sequences as templates and the following primer pairs: SmTK3-TK-pcDNA3B-5′ (5′-GGATCCATGCTCATTGATAAATGGGAAATTCC-3′) + SmTK3-TK-pcDNA3B-3′ (5′-GCGGCCGCCTGGTTGCTCATCTTCACAGA-3′); SmTK6-TK-pcDNA3B-5′ (5′-GGATCCATGTTTGCAATTATCCGTGATAG-3′) + SmTK6-TK-pcDNA3B-3′ (5′-GCGGCCGCCTAAATATTGAGCTTCTGTGTGCG-3′); and SmAbl1-TK-pcDNA3B-5′ (5′-TGGAATTCTATGCCCGAAATTATAATGCGTC-3′) + SmAbl1-TK-pcDNA3B-3′ (5′-GCGGCCGCCTTGTTCCAGTTCCGCA-3′). 5′ and 3′ primers contained, respectively, BamHI and NotI sites that were used to direct the insertion of amplification products into pcDNA3.1B, a T7 promoter-containing plasmid. The resulting SmTK3-TK, SmTK6-TK, and SmAbl1-TK constructs were sequenced confirming their correct ORFs. Plasmids were linearized by PmeI. Capped messenger RNA (cRNA) encoding the different TK domains were synthesized in vitro using the T7 mMessage mMachine kit (Ambion) and analyzed as described previously (21). cRNA preparations were microinjected in Xenopus laevis stage VI oocytes according to a standard protocol (22). Each oocyte was injected with 60 nl (60 ng) of cRNA in the equatorial region and incubated at 19 °C in ND96 medium. After 18 h, GVBD was detected by the appearance of a white spot at the center of the animal pole. Kinase inhibitor studies were performed using herbimycin A (Tocris Bioscience, 10 mm stock solution in DMSO) and imatinib (Alexis Biochemicals, 170 mm stock solution in water). Sets of 10 oocytes freshly injected with SmTK3-TK, SmTK6-TK, or SmAbl1-TK cRNA were placed in ND96 containing different concentrations of herbimycin A (0.0001 to 10 μm final) or imatinib (0.01 to 100 μm final), and GVBD was observed after 18 h. Noninjected oocytes served as negative controls. As positive controls, the natural hormonal stimulus progesterone was used.
Full-length (fl) SmTK6 and SmTK3 were cloned into pcDNA 3.1. SmTK6 plasmids were linearized by PmeI or by EcoRI (position nt 920) to produce cRNA encoding full-length SmTK6 (SmTK6-fl) or only its N-terminal part containing SH3SH2 domains without the TK domain (SmTK6-SH3SH2), respectively.
Dead kinase variants of full-length SmTK6 and SmTK3 (SmTK6-fl-ko and SmTK3-fl-ko) were generated by changing the Mg2+-binding motif DFG present in their kinase domain into a DNA motif, as described previously (22). SmTK6-SmTK3 interaction studies were performed by co-injection of cRNA produced from the different versions of SmTK6 and SmTK3 plasmids.
cRNA was also produced from a pcDNA 3.1 plasmid encoding full-length SmVKR1 (23) rendered constitutively active by an exchange of the Phe-1167 (close to the potential YY1064–1065 autophosphorylation site) to a glutamic acid residue (SmVKR1YYRE) performed by site-directed mutagenesis as described before (24, 25). SmVKR1YYRE-C-term was amplified by PCR from SmVKR1YYRE in pcDNA 3.1 using the primer pair SmVKR1-C-term-5′ (5′-CCCTGCAGTCAAGGTAGAAACGCTAAACTGTTATC-3′; containing a PstI site) and SmVKR1-C-term-3′ (5′-GGGAATTCCGACGTAAACTGAAAGAAATTGAAAATCG-3′; containing an EcoRI site) and subcloned in the T7-containing plasmid PGBKT7 that also contains the Myc tag. SmVKR1-SmTK6 interaction studies were performed by co-injection of cRNA produced from the different versions of SmTK6 and SmVKR1 plasmids.
The expression of proteins in oocytes was confirmed by immunoprecipitation of lysates according to the procedure described previously (22). Following 24 h of expression, oocytes were lysed in buffer (50 mm HEPES, pH 7.4, 500 mm NaCl, 0.05% SDS, 0.5% Triton X-100, 5 mm MgCl2, 1 mg/ml bovine serum albumin, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 10 μg/ml soybean trypsin inhibitor, 10 μg/ml benzamidine, 1 mm PMSF, 1 mm sodium vanadate) and centrifuged at 4 °C for 15 min at 10,000 × g.
Supernatants were incubated with anti-V5 (1:100; Invitrogen), anti-FLAG (1:100; Sigma), or anti-Myc antibodies (1:100; Invitrogen) at 4 °C overnight. Protein A-Sepharose beads (5 mg; Amersham Biosciences) were added for 1 h at 4 °C. Immune complexes were collected by centrifugation, rinsed three times, resuspended in Laemmli sample buffer, and subjected to a 10% SDS-PAGE. Immune complexes were analyzed by Western blotting using anti-V5 or anti-Myc (1:50,000) antibodies and the advanced ECL detection system (Amersham Biosciences).
Transcriptional Analyses
To confirm the transcription of the bait vector transgene SmTK6-SH2 in transformed yeast cells, a 5-ml overnight culture of an appropriate yeast clone was centrifuged. The pellet was washed twice with PBS and frozen in liquid nitrogen. Cells were disrupted by three freeze/thaw cycles (liquid nitrogen, 37 °C water bath); 1 ml of TriFastTM (PeqLab) was added to the lysate, and total RNA was extracted according to the manufacturer's instructions. The synthesis of cDNA was done with 90 ng of total RNA, a primer specific for the SH2 domain of SmTK6 (SmTK6-SH2–3′), and Sensiscript reverse transcriptase (Qiagen). RT-PCR analyses were performed using ¼ of the cDNA as template, FIREPol Taq polymerase (Solis BioDyne) and the primer combination SmTK6-SH2–5′ + SmTK6-SH2–3′.
To investigate the stage-specific transcription of SmTK6 in S. mansoni, RT-PCR analyses were performed with total RNA of adult or larval worms extracted by TriFastTM (PeqLab) following the manufacturer's instructions. Residual DNA was removed by DNase digestion using RNase-free DNase I (Fermentas). cDNA was synthesized with 1 μg of total RNA, the SmTK6 cDNA-specific primer TK6–3′ (5′-GAATTCTTGTTATGCTATCACG-3′), and Superscript II reverse transcriptase (Invitrogen). Subsequent PCRs were performed with of the cDNA as template, FIREPol Taq polymerase (Solis BioDyne), and the following primer combination: TK6–5′ (5′-ATAGTGGGCAAGATGGTGGA-3′) + TK6–3′ (see above; 1 μm each) amplifying a 454-bp product. All PCRs were performed in a final volume of 25 μl. PCR products were separated in 1.5% agarose gels stained with ethidium bromide.
To investigate the stage-specific transcription of SmDLG in S. mansoni, RT-PCR analyses were performed as described above using the primer DLG-5′ (5′-CAAGTACAGGCAGTGCAGGA-3′) + DLG-3′ (5′-GCACATCCAGCAGTTACACG-3′) amplifying a 406-bp product.
In situ hybridizations were done as described elsewhere in detail (20). In short, adult worm pairs were fixed in Bouin's solution (picric acid/acetic acid/formaldehyde; 15:1:5) before embedding in paraplast (Histowax, Reichert-Jung). Sections of 5 μm were generated and incubated in xylol to remove the paraplast. Following re-hydration, proteins were removed by proteinase K treatment (final concentration 1 μg/ml), and the sections were dehydrated. For hybridization, in vitro-generated transcripts were labeled with digoxigenin following the manufacturer's instructions (Roche Applied Science). Labeled sense and antisense transcripts of SmDLG (406 bp; position 974–1379 bp) or SmVKR1 (323 bp; position 3938–4260) were size-controlled by gel electrophoresis. To prove their quality, transcript blots were made to confirm digoxigenin incorporation by alkaline phosphatase-conjugated anti-digoxigenin antibodies, naphthol-AS-phosphate, and Fast Red TR (Sigma). All in situ hybridizations were performed for 16 h at 42 °C. Sections were stringently washed up to 0.5× SSC, and detection was achieved as described for transcript blots.
In Silico Analyses
The following public domain tools were used for sequence analyses: NCBI-BLAST, the Wellcome Trust Sanger Institute S. mansoni OmniBlast server, GeneDB, and Schisto DB. For phylogenetic analyses, the programs ClustalX 2.0 (26) and TreeViewX 0.5.0 were used. For structural analyses SMART (27) was used.
RESULTS
SmTK6 Full-length cDNA Sequence Reveals Src and Abl Characteristics
By Y2H cDNA library screening using the tandem SH2 domain of SmTK4 as bait, the known Src kinase SmTK3 ((15) accession number CAE51198), and the novel kinase SmTK6 (accession number FN397679) were identified as potential upstream binding partners (13). Comparative analyses of the relative binding strengths by β-gal liquid assays showed a weak interaction between SmTK4 and SmTK3 and a strong interaction between SmTK4 and SmTK6. The binding potential of SmTK4 and SmTK6 was finally confirmed by co-immunoprecipitation (13).
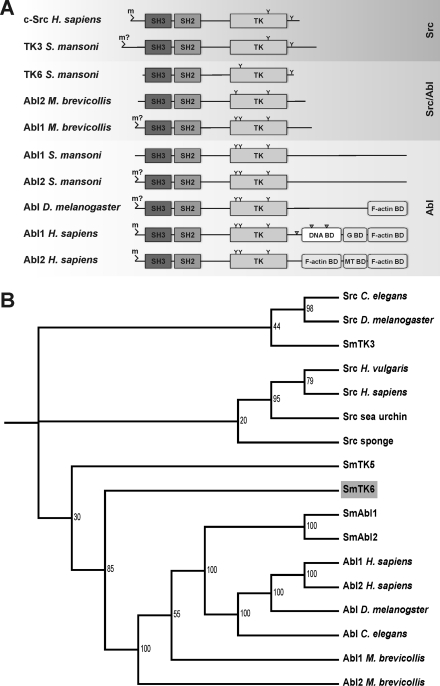
The SmTK6 cDNA has a length of 1698 bp coding for a protein of 565 aa. Domain structures such as SH3 (aa 115–172), SH2 (aa 179–268), and a catalytic TK domain (aa 297–551) are present (supplemental data 1) indicating the similarity of SmTK6 to Src or Abl kinases. An SH4 domain functionally important for intracellular membrane attachment via myristoylation may not exist. Although a typical glycine occurs at position 2 (MGICLC), it is not embedded in a consensus sequence characteristic for myristoylation (MGXXX(S/T)) (28). Because two typical cysteine residues were detected at positions 4 and 6, palmitoylation may be possible instead (29). Compared with other Src kinases, SmTK6 possesses only one of the typical Tyr residues with regulatory function (Tyr-562). It is positioned within the C terminus close to the TK domain at a conserved position compared with Tyr-527 of human c-Src (Fig. 1A and supplemental data 1). Tyr-416 of human Src, which occurs in the C-terminal half of the TK domain, is missing in SmTK6. Instead a serine residue occurs at the corresponding position 447 (supplemental data 1). Within the catalytic TK domain further conserved regions exist, of which two distinguish between serine/threonine kinases and TKs (30). SmTK6 possesses the TK-specific DLAAR(N/D)LRAAN motif of subdomain I (aa 418–423), and the P(I/V)(K/R)W(T/M)APE motif of subdomain II (aa 456–463) supporting the conclusion that SmTK6 is a CTK (supplemental data 1). Src kinase-characteristic sequence motifs also occur within the SH2 and SH3 domains. Within the N-terminal part of the SH3 domain of Src kinases, the consensus sequence ALYDY is located, which concurs well with ALYSY in SmTK6 (aa 119–123). At the C terminus of Src SH3 domains occurs the motif PSNYV matching exactly to that of SmTK6 (aa 164–168). Within the SH2 domains one histidine (aa 233) and three arginine residues occur at conserved positions (aa 186, 206, and 234 in SmTK6; supplemental data 1), which are important for target protein interactions (31). However, SmTK6 also reveals features characteristic for Abl kinases (6). Besides the absence of Tyr-416, a Tyr residue occurs at position 312 of SmTK6. This is one of two conserved Tyr residue typically found in the N-terminal part of Abl TK-domains (Fig. 1A and supplemental data 1). Comparing only the TK domain sequences of Abl and Src kinases by ClustalW analysis indicated that the overall structure of the catalytic domain of SmTK6 is more similar to Abl than to Src kinases (18). Abl kinases of Drosophila or Homo sapiens again possess an elongated C terminus containing various combinations of characteristic binding regions for F- or G-actin, microtubules, or DNA, all of which are absent from SmTK6 as well as a nuclear localization sequence, which was found in the Abl1 kinase of humans (Fig. 1A). Thus, SmTK6 is an Src-biased structural hybrid of both kinase classes representing a potential ancestor of Abl kinases.
FIGURE 1.
A, schematic structures of the functional domains of c-Src from human, the Src kinase SmTK3 and the Src/Abl hybrid kinase SmTK6 from S. mansoni, Abl1 and Abl2 from M. brevicollis, Abl1 and Abl2 from S. mansoni, Abl from Drosophila melanogaster, and Abl1 and Abl2 from human. m, myristoylation site (m?, predicted but not verified myristoylation site); TK, tyrosine kinase domain; G BD, G-actin-binding domain; MT BD, microtubule-binding domain; F-actin BD, F-actin-binding domain; Y, conserved tyrosine phosphorylation site; gray triangle, nuclear location site (NLS). B, dendrogram of the phylogenetic analysis of the SH3-SH2-TK cassette sequences of the Src/Abl hybrid kinase SmTK6, the Src kinases SmTK3 and SmTK5, and the Abl kinases SmAbl1 and SmAbl2 of S. mansoni, as well as other metazoan CTKs using ClustalX and TreeViewX. Bootstrap values are indicated. Sequences were obtained from the National Center for Biotechnology Information using the Entrez Browser (www.ncbi.nlm.nih.gov) and from the M. brevicollis genome website. The corresponding protein accession numbers are as follows: Abl D. melanogaster (protein-tyrosine kinase Abl, D. melanogaster; AAA28934); Abl1 H. sapiens (tyrosine-protein kinase Abl1 isoform b, H. sapiens; NP_009297); Abl2 H. sapiens (tyrosine-protein kinase Abl2 isoform b, H. sapiens; NP_009298); Abl C. elegans (tyrosine-protein kinase Abl-1, C. elegans; P03949); SmAbl1 (Abl protein-tyrosine kinase 1, S. mansoni; CBH50761); SmAbl2 (Abl protein-tyrosine kinase 2, Schistosoma mansoni; CBH50762); Abl1 M. brevicollis (Abl protein kinase 1, M. brevicollis; XP_001742753); Abl2 M. brevicollis (Abl protein kinase 2, M. brevicollis; XP_001746037); Src C. elegans (protein-tyrosine kinase F49B2.5, C. elegans; CAB04427); Src D. melanogaster (Dsrc41, D. melanogaster; BAA07705); Src sponge (tyrosine-protein kinase isoform SRK1, Spongilla lacustris; P42686); Src human (proto-oncogene tyrosine-protein kinase SRC, H. sapiens; NP_005408); Src sea urchin (Src-type protein-tyrosine kinase, Anthocidaris crassispina; BAA33741); Src Hydra (Src-related protein-tyrosine kinase, Hydra vulgaris; AAA29217); SmTK5 (Src/Fyn tyrosine kinase 5, S. mansoni; AAF64151); SmTK3 (Src tyrosine kinase, S. mansoni; CAE51198); and SmTK6 (Src tyrosine kinase, S. mansoni; CAZ50862).
To find additional support for the hybrid character, a phylogenetic analysis was performed using sequence cassettes representing the regulatory regions SH3, SH2, and TK each for comparison. SmTK5, a Fyn kinase-like CTK of S. mansoni (32), was taken as outgroup. This analysis confirmed the Src/Abl intermediate position of SmTK6 supporting its ancestral role as a potential Abl precursor (Fig. 1B).
Inhibitor Studies in Xenopus Oocytes Reveal an Intermediate Src/Abl Kinase Activity of SmTK6
To elucidate whether SmTK6 possesses Src or Abl kinase-like activity, biochemical studies were performed in Xenopus oocytes. As shown before, schistosome kinases can be efficiently expressed in this system, and kinase activities were proven by their capacity to induce GVBD as a resumption of meiosis (13, 21, 22, 25, 33). To be enzymatically active, kinases have to adopt an open conformation, which is usually acquired following interaction with an activated partner molecule such as a receptor. In the absence of an activating partner in Xenopus oocytes, the activity of a schistosome CTK expressed in this system can be achieved if its catalytic TK domain is expressed devoid of regulatory domains responsible for inactive conformation (13). Therefore, cRNA encoding the TK domain of SmTK6 (SmTK6-TK) was injected into Xenopus oocytes. Results showed that the expression of SmTK6-TK protein induced 100% GVBD in oocytes demonstrating the kinase activity of SmTK6-TK (Table 1). This activity was completely abolished when the Src kinase-specific inhibitor herbimycin A was added at a concentration of 10 μm. herbimycin A was shown before to be able to block the activity of the Src kinase SmTK3 and to reduce male-induced mitotic activity in paired females as well as egg production in worms cultured in vitro (16, 34). In the Xenopus oocyte assays, we confirmed that herbimycin A was a potent inhibitor of SmTK3 that completely blocked its activity already at a concentration of 0.01 μm (Table 1). In contrast, the kinase activity of SmAbl1-TK, the TK domain of an Abl kinase homolog from S. mansoni (18), was insensitive to high doses of herbimycin A (100% GVBD at 0.1 μm) and was only partially inhibited at concentrations of 1 μm (80% GVBD) and 10 μm (60% GVBD).
TABLE 1.
Influence of herbimycin A or imatinib on the capacity of the catalytic TK domains of SmTK3, SmTK6, or SmAbl1 expressed in Xenopus oocytes to induce GVBD
SmTK3-TK-induced GVBD (numbers represent % GVBD; mean of two independent experiments) was completely blocked by herbimycin A at a concentration of 0.01 μm. Total inhibition of SmTK6-TK-induced GVBD required 10 μm. At this concentration, SmAbl1-TK was still active (60%). − = not determined.
| Herbimycin A | 0.0001 μm | 0.001 μm | 0.01 μm | 0.1 μm | 1 μm | 10 μm | Control |
|---|---|---|---|---|---|---|---|
| SmTK3-TK | 100 | 80 | 0 | 0 | 0 | 0 | 100 |
| SmTK6-TK | − | − | 100 | 100 | 88 | 0 | 100 |
| SmAbl1-TK | − | − | − | 100 | 80 | 60 | 100 |
In an inhibitor swap-like experiment, we also compared the effect of the Abl inhibitor imatinib on the capacity of SmTK6-TK and SmAbl1-TK to induce GVBD (Table 2). This specific inhibitor has been successfully used in human cancer therapy (Gleevec, Novartis) to treat chronic myelogenous leukemia, which is caused by deregulation of the c-Abl kinase (35, 36). Furthermore, in vitro culture studies with adult schistosomes demonstrated recently that imatinib significantly affected schistosome morphology and physiology. Besides negative effects on gonad development and pairing stability, this inhibitor caused pathological alterations of the gastrodermis leading to parasite death (18). Whereas in Xenopus oocytes SmAbl1-TK activity was completely blocked at a concentration of 1 μm, the total inhibition of SmTK6-TK activity required 100 μm imatinib, a concentration at which SmTK3-TK activity was only slightly reduced (by 10%; Table 2).
TABLE 2.
Symbolism is as in Table 1
Imatinib completely blocked SmAbl1-TK at 1 μm. At this concentration, the activities of SmTK3-TK or SmTK6-TK were not affected. Although SmTK3-TK was still active (90%) at 100 μm imatinib, SmTK6-TK was totally inhibited at this concentration. − = not determined.
| Imatinib | 0.01 μm | 0.1 μm | 1 μm | 10 μm | 100 μm | Control |
|---|---|---|---|---|---|---|
| SmTK3-TK | − | − | 100 | 100 | 90 | 100 |
| SmTK6-TK | − | − | 100 | 80 | 0 | 100 |
| SmAbl1-TK | 100 | 90 | 0 | 0 | 0 | 100 |
As expected, these results confirmed the biochemical nature of SmTK3 as an Src-like kinase and also the Abl kinase-like nature of SmAbl1. More importantly, these data demonstrated an inhibition profile of SmTK6 being intermediate to that of Src and Abl kinases.
Transmembrane Receptors, an Src Kinase, and a Tumor Suppressor Molecule Are Potential Upstream Binding Partners of SmTK6
Signaling molecules of the Src and Abl kinase families each possess one SH2 domain, which is known to interact with upstream binding partners in signaling cascades. Using a bait construct expressing the SH2 domain of SmTK6, the S. mansoni adult stage Y2H cDNA library (20) was screened. Expression of the bait was confirmed at the transcriptional level by RT-PCR analyses using RNA extracts from transformed yeast cells. Screening resulted in the identification of 77 initial prey clones, which underwent growth selection and β-gal filter assays (color selection) reducing the number to 29 clones. The isolation of the prey plasmids succeeded for 19 of these. Their inserts represented partial sequences of protein coding genes for which full-length (fl) cDNA sequences were detected in the genome data set of the S. mansoni sequencing project (37) or in the NCBI database (www.ncbi.nlm.nih.gov) using the obtained sequences for BlastX analyses. The results showed that four clone groups (Table 3) were represented consisting of a transmembrane mucin homology group (named SmTmMuc1, Table 3, part A), the Src kinase SmTK3 (part B), a tumor suppressor protein with homology to Discs-large (SmDLG, part C), and proteins with no significant homology (part D).
TABLE 3.
Upstream binding partners of SmTK6 identified by Y2H analyses
Following sequencing of the 19 clones obtained from library screening, BlastX analysis revealed that they represented four groups (A–D) with homology to mucin (group A), the schistosome Src kinase SmTK3 (group B), a cell polarity protein with homology to Discs-large (group C), and proteins with no significant homology (group D). Appropriate full-length cDNA sequences were identified in the S. mansoni genome data set (see Ref. 36) or in the NCBI database (www.ncbi.nlm.nih.gov). Accession and Smp numbers, sizes of the full-length (fl) sequences, the appropriate e-values, and the clone numbers are given.
| Clone group | Homology (accession/Smp number) | Size (bp) | e-value | Clone no. |
|---|---|---|---|---|
| A | Mucin homolog (H. sapiens, Q02817); fl: XP_002578516, Smp_161910 | 9849 | 1e−48 | 1, 6, 9, 10, 18, 36, 38, 40, 43, 45, 54, 59, 63 |
| B | SmTK3 (Src kinase) (S. mansoni, CAE51198); fl: XP_002576744, Smp_151300 | 1944 | 0.0 | 50, 56, 58 |
| C | Cell polarity protein/Discs-large homolog (H. sapiens, NP_001136172); fl: XP_002579880, Smp_170290 | 3432 | 2e−72 | 41 |
| D | No significant homology | 17, 66 |
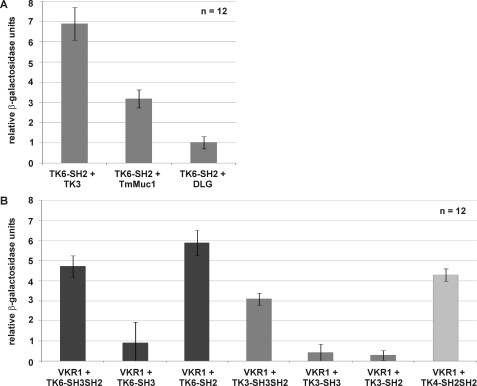
To confirm interactions of potential binding partners with known identity (SmTK3, SmDLG, and SmTmMuc1) and to determine their relative binding strengths in a comparative approach, yeast cells (AH109) were transformed with appropriate prey plasmids together with the bait construct SmTK6-SH2 pBridge. After transformation, all yeast clones survived growth and color selection. To quantify the relative strengths of interaction, β-gal liquid assays were performed. The results again confirmed the observed interactions with SmTK6 and demonstrated the strongest affinity between the SmTK6 SH2 domain and SmTK3. The interactions between the SmTK6 SH2 domain and SmDLG or SmTmMuc1 were considerably weaker (Fig. 2A).
FIGURE 2.
A, yeast cells (strain AH109) were re-transformed with one representative prey clone of each group together with the bait SmTK6-SH2 pBridge, and the relative β-gal activity was measured. Tested clones were (from left to right) as follows: SmTK3 (Src kinase), the mucin homolog SmTmMuc1, and the Discs large homolog SmDiscs large. The statistical evaluation of 12 independent measurements of β-gal activity (nbiol = 2, each with ntech = 6) is shown (error bars are indicated). B, yeast cells (strain AH109) were re-transformed individually with the prey plasmid SmVKR1-C-term pACT2 together with the baits (from left to right) SmTK6-SH3SH2 pBridge, SmTK6-SH3 pBridge, SmTK6-SH2 pBridge, SmTK3-SH3SH2 pBridge, SmTK3-SH3 pBridge, SmTK3-SH2 pBridge, or SmTK4-SH2SH2 pBridge. The relative β-gal activity was measured. The statistical evaluation of six independent measurements (nbiol = 2, each with ntech = 6) is shown (error bars are indicated). Dark gray columns represent the involvement of SmTK6 constructs; gray columns represent SmTK3 constructs, and the light gray column represents the SmTK4 construct.
Literature data have indicated that Src kinases can act in concert with Syk kinase to participate in membrane receptor complexes, which among others contain RTKs (38). SmVKR1 of S. mansoni is an RTK and was shown to be expressed in the ovary of females (23), thus co-localizing with SmTK6 and SmTK4 (13, 17). Therefore, we investigated whether SmVKR1 may interact with these CTKs. To this end, direct binding studies were performed in the Y2H system with the intracellular part of SmVKR1 and the protein interaction domains of SmTK6, SmTK3, or SmTK4. Yeast cells (AH109) were transformed with the prey construct SmVKR1 C-term pACT2 and with bait plasmids containing both or individual SH3 and SH2 domains of SmTK6 or SmTK3 or the tandem SH2 domain of SmTK4. Following transformation, all yeast clones survived growth and color selections indicating that all three kinases were able to bind by their SH2/SH3 domains to the intracellular part of SmVKR1. This was confirmed by β-gal liquid assays, which also provided a first hint to the relative strengths of the observed interactions demonstrating the strongest interaction of SmTK6 with the C terminus of SmVKR1 (Fig. 2B). As expected, the interaction of the SH2 domain of SmTK6 was stronger compared with the interaction of its SH3 domain or the combined SH3SH2 domains. In contrast, the combined SH3SH2 domains of SmTK3 interacted stronger with the intracellular part of SmVKR1 than the individual SH2 or SH3 domains of SmTK3. Finally, the interaction of the tandem SH2 domain of SmTK4 to SmVKR1 was stronger than the interaction of SmTK3-SH3SH2 but weaker than the interaction of SmTK6-SH2 with SmVKR1.
Co-immunoprecipitation Experiments Confirm SmTK6-SmTK3 and SmTK6-SmVKR1 Interactions
Previous studies showed that SmTK3 and SmVKR1 are expressed in the gonads of schistosomes co-localizing in the ovary (15, 23). Because SmTK4 and SmTK6 are also transcribed in the ovary of females, and because by Y2H library screening SmTK6 was found as a binding partner, whose interaction with SmTK4 was confirmed by co-immunoprecipitation experiments (13), a kinase complex acting in the gonads was first hypothesized (13, 14).
To provide further evidence for such a complex, co-immunoprecipitation experiments were performed with the strongest interaction partners SmTK3 and SmVKR1. To this end the Xenopus oocyte expression system was used again.
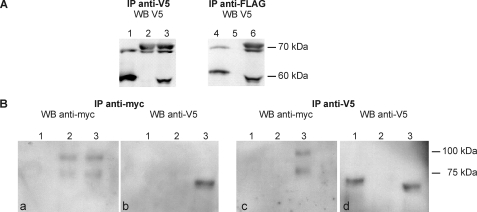
Lysates of oocytes expressing FLAG/V5-tagged SmTK6-fl and/or V5-tagged SmTK3-fl were immunoprecipitated by anti-V5 antibodies and then analyzed by Western blot using the same serum, which confirmed the expression of both proteins (Fig. 3A, lanes 1–3). Anti-FLAG antibodies were then used to immunoprecipitate selectively SmTK6-fl (Fig. 3A, lane 4) but not SmTK3 (Fig. 3A, lane 5). When SmTK6 and SmTK3 were co-expressed, anti-FLAG antibodies immunoprecipitated an additional V5-derived band representing SmTK3 (Fig. 3A, lane 6).
FIGURE 3.
A, tagged full-length variants of SmTK6 and SmTK3 were expressed alone or together in Xenopus oocytes (lanes 1 and 4, FLAG/V5 SmTK6-fl; lanes 2 and 5, V5-SmTK3-fl; lanes 3 and 6, FLAG/V5-SmTK6-fl + V5-SmTK3-fl), immunoprecipitated (IP) by anti-V5 (left) or anti-FLAG antibodies (right), and then analyzed by Western blot (WB) using anti-V5 antibodies. SmTK6 has been detected as expected at 55 kDa and SmTK3 with the expected size of 70 kDa. B, V5-SmTK6-fl and Myc-tagged SmVKR1YYRE-C-term were expressed alone or together in Xenopus oocytes (lane 1, V5-SmTK6-fl; lane 2, myc-SmVKR1YYRE-C-term; lane 3, V5-SmTK6-fl + myc-SmVKR1YYRE-C-term) imunoprecipitated (IP) by anti-Myc (left pair) or anti-V5 (right pair) antibodies, and then analyzed by Western blot using the same antibodies. SmTK6 is expected at 55 kDa and SmVKR1-C-term at 70 kDa (the additional upper band is supposed to represent post-translational modifications of VKR1).
For interaction analyses of SmTK6 and SmVKR1, FLAG/V5-tagged SmTK6-fl and the active kinase of SmVKR1 (Myc-tagged SmVKR1YYRE-C-term) constructs were co-expressed in oocytes. SmVKR1 was detected in anti-Myc immunoprecipitates when injected alone or with SmTK6 following anti-Myc Western blotting (Fig. 3B, panel a, lanes 2 and 3). Similarly, SmTK6 was detected in anti-V5 immunoprecipitates when injected without or with SmVKR1 (Fig. 3B, panel d, lanes 1 and 3) confirming the correct expression of both proteins in oocytes. Results of the analyses of oocyte extracts containing both SmVKR1 and TK6 showed that V5-tagged SmTK6 was contained in anti-Myc precipitates (Fig. 3B, panel b, lane 3) and, inversely, Myc-tagged SmVKR1 was present in anti-V5 precipitates (Fig. 3B, panel c, lane 3) demonstrating the interaction between the two molecules. In summary, these co-immunoprecipitation experiments confirmed the physical interactions of SmTK6 with both SmTK3 and SmVKR1.
SmTK6 Is Activated by SmVKR1 or SmTK3 and Induces GVBD in Xenopus Oocytes
To confirm that SmVKR1 not only interacts with but also activates SmTK6, cRNAs encoding the full-length version of SmTK6 (SmTK6-fl) or its SH2-SH3 protein interaction domains (SmTK6-SH3SH2) were co-injected into Xenopus oocytes with SmVKR1YYRE, the constitutively active kinase mutant of SmVKR1. Comparative GVBD assays were performed (Table 4). As expected, the expression of SmTK6-fl alone did not lead to GVBD because of its inactive kinase conformation. SmTK6-SH2SH3 alone was also unable to induce GVBD, in contrast to the SmTK6-TK catalytic domain, which induced 100% GVBD (Tables 1 and 2). The expression of SmVKR1YYRE alone also led to GVBD, whereas wild-type SmVKR1 in the absence of ligand had no effect as shown previously (25). The results showed that the co-expression of SmTK6-fl with SmVKR1YYRE did not affect GVBD induced by the SmVKR1 active kinase. However, when the SH2SH3 domains of SmTK6 were co-expressed with SmVKR1YYRE, GVBD was totally inhibited, suggesting that binding of protein interaction domains of SmTK6 to SmVKR1 could specifically compete with partners found by SmVKR1 in oocytes preventing its GVBD-inductive potential. Also, the demonstration that a “kinase-dead” variant of SmTK6-fl (SmTK6-fl-ko) was able to block the SmVKR1YYRE-induced GVBD in oocytes confirmed that molecular interactions effectively occur between SmTK6 and SmVKR1, leading possibly to SmTK6 kinase activation, as a step of the signaling cascade required for oocyte maturation.
TABLE 4.
SmTK6 activation due to SmVKR1 interaction in Xenopus GVBD assays
cRNAs of SmTK6 full-length (fl), protein-binding domains only (SH3SH2), or a kinase-dead variant of the full-length SmTK6 (fl-ko) were co-injected with cRNAs of the wild-type form of SmVKR1 (WT) or of its constitutively active mutant (YYRE) in oocytes. Numbers represent % GVBD (mean of two independent experiments).
| None | SmVKR1WT | SmVKR1YYRE | |
|---|---|---|---|
| None | 0 | 80 (±10) | |
| SmTK6-fl | 0 | 0 | 85 (±5) |
| SmTK6-SH3SH2 | 0 | 0 | 0 |
| SmTK6-fl-ko | 0 | 0 | 0 |
Because SmTK3 represented the other potent interaction partner of SmTK6, we investigated its potential to alternatively induce SmTK6 catalytic activity too. SmTK6-fl or SmTK3-fl alone were not able to induce GVBD in Xenopus oocytes (Table 5), whereas their co-expression elicited GVBD. However, the co-expression of SmTK3-fl with the kinase-dead variant SmTK6-fl-ko or with the SmTK6-SH3SH2 domains had no inductive effects on oocyte maturation, indicating that the full-length version of SmTK6 is required for activation. In contrast, it appeared that the presence of a full-length and enzymatically active version of SmTK3 was not required to obtain GVBD in this system because the co-expression of a kinase-dead variant of SmTK3 (SmTK3-fl-ko) or of only the TK domain of SmTK3 with SmTK6-fl induced oocyte maturation (Table 5). These results indicated that SmTK6 gets activated independently of the kinase activity of SmTK3. Indeed, the ability of the dead kinase SmTK3-fl-ko to activate SmTK6-fl was corroborated by the observation that herbimycin A (used at a concentration already shown to block SmTK3 kinase activity completely; Tables 1 and 2) did not inhibit GVBD induced by the co-expression of SmTK3-TK with SmTK6-fl. Finally, these results provided evidence that the kinase potential of SmTK6 leading to GVBD is positively controlled by its interaction with SmTK3 (Table 5).
TABLE 5.
SmTK6 activation due to SmTK3 interaction in Xenopus GVBD assays
cRNAs of SmTK6 full-length (fl) or protein-binding domains only (SH3SH2) were co-injected with cRNAs of SmTK3 full-length (fl) wild-type form or of its kinase-dead mutant (ko) or of SmTK3-TK domain in oocytes. The given % GVBD is the mean of two independent experiments.
| SmTK6 variant | SmTK3 variant | GVBD |
|---|---|---|
| % | ||
| SmTK6-fl | 0 | |
| SmTK3-fl | 0 | |
| SmTK6-fl | SmTK3-fl | 80 |
| SmTK6-fl-ko | SmTK3-fl | 0 |
| SmTK6-SH3SH2 | SmTK3-fl | 0 |
| SmTK6-fl | SmTK3-fl-ko | 85 (±5) |
| SmTK6-fl-ko | SmTK3-fl-ko | 0 |
| SmTK6-fl | SmTK3-TK | 90 |
| SmTK6-fl | SmTK3-TK + HerbAa | 100 |
a Herbimycin A (HerbA) was added to a concentration of 0.01 μm to block the catalytic activity of SmTK3.
Transcriptional Analyses Demonstrate Similar Activity Profiles of Interaction Partners
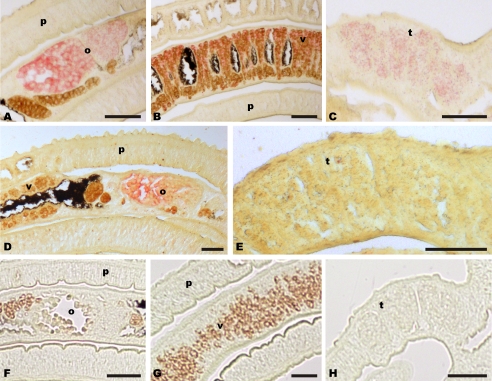
RT-PCR analyses showed that SmTK6 transcription observed in males and females corresponded to the transcriptional profile of SmDLG; however, differences were observed for the larval stages because SmDLG transcripts were detected only in miracidia and not in cercariae (supplemental data 2). In contrast, SmTmMuc1 was found to be transcribed in the adult stages as well as in both larval stages,3 which corresponded also to the transcriptional profiles of SmTK3 (15) and SmTK4 (17). Co-localization of SmTK6 with SmTK4 transcripts in the testes and the ovary of adult schistosomes had already been demonstrated before (13). As shown by in situ hybridization, SmDLG co-localized with SmTK6 in the gonad tissues of adult worms (Fig. 4, A–C). This applies also to SmTmMuc1 transcripts whose occurrence was observed in the gonads and also in other tissues.3 In particular, the transcription pattern in the reproductive organs of SmDLG and SmTmMuc1 corresponded to that of SmTK6, and there is additional co-localization with SmTK4 in the ovary and in the testes. Finally, transcripts of SmVKR1 used as positive control were detected as expected exclusively in the ovary of females (Fig. 4, D and E), which corresponded to previous data (22). Sections of control worms hybridized with sense transcripts did not show any signal. The localization results indicated that the ovary is the organ where SmTK6 and SmVKR1 co-localize with all other molecules investigated in this study and also with SmTK3 and SmTK4.
FIGURE 4.
Nonconserved regions of SmDLG (A–C) and SmVKR1 (D and E) served as templates to synthesize digoxigenin-labeled antisense transcripts used as probes for in situ hybridization. SmVKR1 signals were exclusively detected in the ovary (o), whereas SmDLG signals were detected in the ovary and the vitellarium (v) of the female as well as in the testes (t) of the male. p, parenchyma. For control, a digoxigenin-labeled sense probe was used (F–H). (Scale bar, 50 μm.)
DISCUSSION
Structural and phylogenetic analyses performed in this study provided conclusive evidence that SmTK6 represents an exceptional hybrid kinase taking an intermediate evolutionary position between Src and Abl kinases by being the first hybrid kinase detected in a helminth. Members of both kinase families have been found in the majority of metazoans, which suggests that their structure and function were fixed early in evolution. Vertebrate genomes encode two closely related paralogs, Abl1 and Abl2, which may have originated from a gene duplication event. Invertebrate metazoans such as Caenorhabditis elegans or Drosophila possess a single Abl gene, which shows strong conservation through the SH3-SH2-TK cassette and an elongated C terminus with an actin-binding domain (6). The choanoflagellate Monosiga brevicollis, a unicellular protist, encodes two Abl kinases with a shortened C-terminal end without an actin-binding domain. This suggested an early origin for Abl kinases, with the addition of extended C termini during the metazoan radiation (6). In conformity with this hypothesis, schistosomes and perhaps other trematodes seem to be gap-filling in evolutionary terms by possessing two Abl genes with elongated C termini. But none of these contains a conserved actin-binding domain (Fig. 1A). SmTK6 has no actin-binding domain as well and exhibits the shortest C terminus found so far for Src or Abl kinases. Within its C terminus, the Src characteristic Tyr-527 occurs, but the regulatory Tyr-416 is missing. Instead, one of two Abl kinase-specific Tyr residues occur (Fig. 1A). This finding and the results of the phylogenetic analyses suggested a hybrid character of this unusual kinase. Functional evidence in support of this hypothesis was obtained by GVBD assays in Xenopus oocytes expressing the catalytic TK domains of SmTK6 and other schistosome kinases representing members of the Src (SmTK3) or Abl (SmAbl1) kinase families (15, 18). Using inhibitors specific for each of these CTK families demonstrated the hybrid character also at the biochemical level. SmTK6-TK-induced GVBD was inhibited by the Src kinase inhibitor herbimycin A at a ×1000 higher concentration than that needed to inhibit SmTK3-TK-induced GVBD. The Abl kinase inhibitor imatinib was also able to completely block SmTK6-TK activity but at a ×100 higher concentration than that needed to inhibit SmAbl1-TK-induced GVBD. Thus, the inhibition profile of SmTK6 was found to be intermediate between Src and Abl kinases. In a previous study, it was shown that 21-aa residues of the human Abl kinase interact with imatinib (39). Of these, 18 are conserved in SmAbl1 according to our analysis but only 15 in SmTK6 (18). This difference possibly explains the reduced inhibitory effect of imatinib on SmTK6 compared with SmAbl1. Among the differences is an amino acid substitution next to the conserved DFG motif within the catalytic TK domain. In contrast to Abl kinases of Drosophila, C. elegans, or H. sapiens, which exhibit a conserved ADFGL (aa 381–385) sequence motif, SmTK6 has SDFGL instead (aa 435–438 in SmTK6; supplemental data 1). Because high affinity binding of imatinib requires a flipped DFG motif (40), the nonsynonymous Ala/Ser substitution may contribute to this conformational change and negatively influence inhibitor binding.
As results from Y2H library screening schistosome homologs of the known Src kinase SmTK3 (15), the tumor suppressor protein DLG and a novel transmembrane mucin were found as potential interaction partners of SmTK6. RT-PCR analyses showed overlapping transcriptional profiles of all genes in adult stages, and in situ hybridizations finally demonstrated co-localization in the gonads of adults. Interactions of SmTK6 were also found with the co-localizing SmVKR1 (23). This RTK may not have been represented in or picked-up from the library because of a potential competition between membrane localization signals within its sequence and the nuclear location site sequences of the prey vector destined to ensure nuclear interaction and reporter gene induction.
DLG is a tumor suppressor protein and a prototype of a growing family of proteins collectively termed membrane-associated guanylate kinase homologs. Genetic studies in Drosophila revealed that three tumor suppressors, DLG, Scribble, and Lethal giant larvae, co-localized to the basolateral region of epithelial cells cooperatively regulating cell polarity, junction formation, and cell growth during oogenesis (41). Studies in vertebrates and C. elegans demonstrated the evolutionary conservation of some of their functions (42), and new evidence has indicated a role of membrane-associated guanylate kinase homologs in asymmetric cell division (43), a characteristic feature of germ cells (44). A presumptive interaction of Src kinases and PDZ (Postsynaptic density SD95/SAP90, DLG, Zonula occludentes 1) domain-containing proteins such as DLG was shown recently (45). Because the partial SmDLG clone contained a PDZ domain, interaction with SmTK6 may have been mediated by this domain. Recently, we found an S. mansoni Lethal giant larvae homolog (SmLGL) whose transcripts were also localized in the reproductive organs.4 Because two schistosome homologs of Scribble exist in the genome (Smp_180220.2; Smp_104030.2), all members of a cooperatively acting molecular network are present in this parasite that have been shown before in Drosophila to play roles during oogenesis (41).
As a group of high molecular weight glycoproteins, mucins are divided into secreted and membrane-bound forms containing a single transmembrane domain (46). Members of the latter class are involved in cellular signaling events, and they contribute to carcinogenesis (47). Among others, transmembrane mucins such as human MUC1 have the potential to interact with RTKs such as EGF receptors (48). The co-localization of SmTmMuc1 with SmTK6 and other schistosome kinases, including SmVKR1, suggests a role in signal transduction processes in the gonads of schistosomes in concert with further signaling molecules.3
Src kinases are known to be involved in signaling pathways regulating cell proliferation and differentiation (1–3). Often they are parts of large complexes consisting of different cellular, membrane-associated, and transmembrane signaling proteins. SmVKR1 (previously called SmRTK-1) was described as an unusual schistosome RTK composed of an extracellular Venus Flytrap module (a ligand-binding domain in class 3 G-protein-coupled receptors) linked by a single transmembrane domain to a TK domain, which displayed similarity to that of insulin receptors (23). Venus kinase receptors have been found in different invertebrates, particularly in insects (24). As in schistosomes, Venus kinase receptors are mainly expressed in female gonads, indicating a putative function of these receptors in reproduction and/or development. It has been hypothesized that SmVKR1 might be involved in the recognition of a male pheromone signal necessary for the development and maturation of the ovary because it possesses a Venus Flytrap module, also present in mammalian pheromone receptors (14, 23, 49). SmVKR1 transcripts were predominantly found in mature oocytes of female schistosomes by in situ hybridization, which coincided with the transcript localizations of SmTK4 (17), SmTK3 (15), and SmTK6 (13). In addition to the SmTK4-SmTK6 binding confirmed previously (13), we showed in this study by co-immunoprecipitation experiments that SmTK6 interacts with SmTK3 as well as with SmVKR1. In addition, GVBD assays in Xenopus oocytes demonstrated that SmTK6 interacted with and is likely activated by catalytically active SmVKR1. Furthermore, we showed that SmTK3 is also able to activate SmTK6. For activation, binding to SmTK3 alone was sufficient and independent from the catalytic potential of SmTK3. We assume that SmTK6 changes into an open conformation upon SmTK3 binding as a prerequisite to become active inducing GVBD in Xenopus oocytes. A question still to solve in further studies is whether SmTK6 gets phosphorylated and finally activated by SmVKR1 and whether SmTK3 in such a scenario plays a chaperone-like role supporting the conformational change of SmTK6 for further catalytic interactions within the postulated multikinase complex.
Another attractive goal of this study was to knock down SmTK6 by RNA interference (RNAi) to study the phenotype among others by confocal laser scanning microscopy. This approach has been successfully applied before to characterize schistosome kinases such as SmTK4 (13). Although standard protocols (dsRNA) (13, 50) as well as novel approaches using different sets of specifically designed siRNAs5 were applied by electroporation in adult schistosomes maintained in vitro, we obtained neither knockdown effects at the transcriptional level (determined by qPCR) nor phenotypic changes (as determined by confocal laser scanning microscopy (13)). SmTK6 may belong to the group of genes, which according to recent studies in this field were described as nonknockable genes (50).
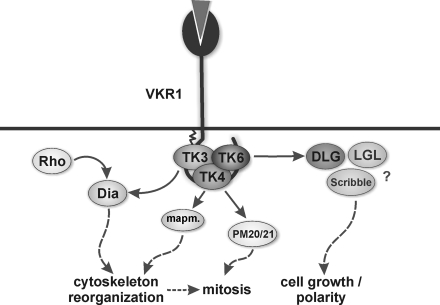
With respect to their interactions and co-localizations in the reproductive organs, SmTK6, SmVKR1, SmTK3, SmTK4, and SmDLG seem to be parts of a complex scenario (Fig. 5). In previous studies, evidence was obtained that SmTK3 interacts with SmTK4 and that both may be members of a CTK complex (14). Together with the Rho-GTPase SmRho1 and the diaphanous homolog SmDia, SmTK3 was suggested to be part of integrating RTK- and G-protein-coupled receptor signaling pathways, which organize the actin cytoskeleton within the gonads of schistosomes (20). Homologs of a MAPK-activating protein (PM20/21) and mapmodulin were found before as downstream partners of SmTK4 and with SmTK6 as its strongest upstream partner (13). CTK members of this multikinase complex such as SmTK6 are probably controlled by SmVKR1 (Fig. 5). SmDLG as a further binding partner of SmTK6 may become activated upon complex formation. Subsequently, SmDLG may interact with other membrane-associated guanylate kinase homologs such as SmLGL and Scribble to control processes of cell growth and/or cell polarity.
FIGURE 5.
In S. mansoni the CTKs SmTK3, SmTK4, and SmTK6 may be members of a trimeric complex, which interacts with the RTK SmVKR1. Results of a previous study had already indicated that SmTK3 also interacted with the diaphanous homolog SmDia, which is a binding partner of the Rho-GTPase SmRho1. Both SmDia and SmRho1 were suggested to be part of cooperative RTK and G-protein-coupled receptor signaling pathways integrating at SmDia to organize the actin cytoskeleton within the gonads of schistosomes (20). As downstream partners of SmTK4, MAPK-activating protein (PM20/21) and mapmodulin were found, which may be involved in cytoskeleton reorganization and mitosis (13). SmDLG as a binding partner of SmTK6 may become activated upon complex formation and may subsequently interact with SmLGL and Scribble to control processes of cell growth and/or cell polarity.
Elucidating cellular processes leading to gonad development in schistosomes along with the increasing knowledge of molecules involved will help to find novel strategies to fight this blood fluke. To this end, kinases represent interesting targets to identify inhibitors affecting developmental processes. Such inhibitors could already represent approved drugs (18), or they can serve at least as lead structures for drug design or vaccination (51). This is an urgent need in the light of the fact, that praziquantel is the only widely used drug to treat schistosomiasis, thus evoking the fear of emerging resistance (52, 53).
Supplementary Material
Acknowledgments
We acknowledge the excellent technical assistance of Gabriele Lang, Brigitte Hoffmann, Christine Henrich, and Arlette Lescuyer. We thank the Wellcome Trust Sanger Institute for providing schistosome genome data.
This work was supported by Deutsche Forschungsgemeinschaft Grant GR1549/8-2, Bill and Melinda Gates Foundation Grant OPP1024324 through the Grand Challenges Exploration Initiative, by INSERM, CNRS, Universite de Lille 1-Lille 2, and by the H.-J. and G. Engemann Foundation (to S. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental data 1 and 2.
S. Hahnel and C. G. Grevelding, manuscript in preparation.
C. Buro, S. Beckmann, and C. G. Grevelding, manuscript in preparation.
S. Beckmann, T. Quack, G. Lang, and C. G. Grevelding, manuscript in preparation.
- CTK
- cellular protein-tyrosine kinase
- RTK
- receptor tyrosine kinase
- GVBD
- germinal vesicle breakdown
- SH
- Src homology
- TK
- tyrosine kinase
- aa
- amino acid
- DLG
- Discs-large homolog
- Y2H
- yeast two-hybrid.
REFERENCES
- 1. Hunter T. (2009) Curr. Opin. Cell Biol. 21, 140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bromann P. A., Korkaya H., Courtneidge S. A. (2004) Oncogene 23, 7957–7968 [DOI] [PubMed] [Google Scholar]
- 3. Lemmon M. A., Schlessinger J. (2010) Cell 141, 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Takahashi A., Obata Y., Fukumoto Y., Nakayama Y., Kasahara K., Kuga T., Higashiyama Y., Saito T., Yokoyama K. K., Yamaguchi N. (2009) Exp. Cell Res. 315, 1117–1141 [DOI] [PubMed] [Google Scholar]
- 5. Hubbard S. R., Till J. H. (2000) Annu. Rev. Biochem. 69, 373–398 [DOI] [PubMed] [Google Scholar]
- 6. Colicelli J. (2010) Sci. Signal. 3, re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chitsulo L., Loverde P., Engels D. (2004) Nat. Rev. Microbiol. 2, 12–13 [DOI] [PubMed] [Google Scholar]
- 8. Quack T., Beckmann S., Grevelding C. G. (2006) Berl. Munch. Tierarztl. Wochenschr. 119, 365–372 [PubMed] [Google Scholar]
- 9. Ross A. G., Bartley P. B., Sleigh A. C., Olds G. R., Li Y., Williams G. M., McManus D. P. (2002) N. Engl. J. Med. 346, 1212–1220 [DOI] [PubMed] [Google Scholar]
- 10. Kunz W. (2001) Trends Parasitol. 17, 227–231 [DOI] [PubMed] [Google Scholar]
- 11. LoVerde P. T., Niles E. G., Osman A., Wu W. (2004) Can. J. Zool. 82, 357–374 [Google Scholar]
- 12. Grevelding C. G. (2004) Curr. Biol. 14, R545. [DOI] [PubMed] [Google Scholar]
- 13. Beckmann S., Buro C., Dissous C., Hirzmann J., Grevelding C. G. (2010) PLoS Pathog. 6, e1000769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beckmann S., Quack T., Burmeister C., Buro C., Long T., Dissous C., Grevelding C. G. (2010) Parasitology 137, 497–520 [DOI] [PubMed] [Google Scholar]
- 15. Kapp K., Knobloch J., Schüssler P., Sroka S., Lammers R., Kunz W., Grevelding C. G. (2004) Mol. Biochem. Parasitol. 138, 171–182 [DOI] [PubMed] [Google Scholar]
- 16. Knobloch J., Kunz W., Grevelding C. G. (2006) Int. J. Parasitol. 36, 1261–1272 [DOI] [PubMed] [Google Scholar]
- 17. Knobloch J., Winnen R., Quack M., Kunz W., Grevelding C. G. (2002) Gene 294, 87–97 [DOI] [PubMed] [Google Scholar]
- 18. Beckmann S., Grevelding C. G. (2010) Int. J. Parasitol. 40, 521–526 [DOI] [PubMed] [Google Scholar]
- 19. Grevelding C. G. (1995) Mol. Biochem. Parasitol. 71, 269–272 [DOI] [PubMed] [Google Scholar]
- 20. Quack T., Knobloch J., Beckmann S., Vicogne J., Dissous C., Grevelding C. G. (2009) PloS One 4, e6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Long T., Cailliau K., Beckmann S., Browaeys E., Trolet J., Grevelding C. G., Dissous C. (2010) Int. J. Parasitol. 40, 1075–1086 [DOI] [PubMed] [Google Scholar]
- 22. Vicogne J., Cailliau K., Tulasne D., Browaeys E., Yan Y. T., Fafeur V., Vilain J. P., Legrand D., Trolet J., Dissous C. (2004) J. Biol. Chem. 279, 37407–37414 [DOI] [PubMed] [Google Scholar]
- 23. Vicogne J., Pin J. P., Lardans V., Capron M., Noël C., Dissous C. (2003) Mol. Biochem. Parasitol. 126, 51–62 [DOI] [PubMed] [Google Scholar]
- 24. Ahier A., Rondard P., Gouignard N., Khayath N., Huang S., Trolet J., Donoghue D. J., Gauthier M., Pin J. P., Dissous C. (2009) PLoS One 4, e5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gouignard N., Vanderstraete M, Cailliau K., Lescuyer A., Browaeys E., Dissous C. (2011) Exp. Parasitol., in press [DOI] [PubMed] [Google Scholar]
- 26. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 27. Schultz J., Milpetz F., Bork P., Ponting C. P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Resh M. D. (1994) Cell 76, 411–413 [DOI] [PubMed] [Google Scholar]
- 29. Resh M. D. (2006) Sci. STKE 2006, re14. [DOI] [PubMed] [Google Scholar]
- 30. Hanks S. K., Quinn A. M., Hunter T. (1988) Science 241, 42–52 [DOI] [PubMed] [Google Scholar]
- 31. Overduin M., Mayer B., Rios C. B., Baltimore D., Cowburn D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 11673–11677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kapp K., Schüssler P., Kunz W., Grevelding C. G. (2001) Parasitology 122, 317–327 [DOI] [PubMed] [Google Scholar]
- 33. Yan Y., Tulasne D., Browaeys E., Cailliau K., Khayath N., Pierce R. J., Trolet J., Fafeur V., Ben Younes A., Dissous C. (2007) Int. J. Parasitol. 37, 1539–1550 [DOI] [PubMed] [Google Scholar]
- 34. Knobloch J., Beckmann S., Burmeister C., Quack T., Grevelding C. G. (2007) Exp. Parasitol. 117, 318–336 [DOI] [PubMed] [Google Scholar]
- 35. Manley P. W., Cowan-Jacob S. W., Buchdunger E., Fabbro D., Fendrich G., Furet P., Meyer T., Zimmermann J. (2002) Eur. J. Cancer 38, S19–S27 [DOI] [PubMed] [Google Scholar]
- 36. Larson R. A., Druker B. J., Guilhot F., O'Brien S. G., Riviere G. J., Krahnke T., Gathmann I., Wang Y. (2008) Blood 111, 4022–4028 [DOI] [PubMed] [Google Scholar]
- 37. Berriman M., Haas B. J., LoVerde P. T., Wilson R. A., Dillon G. P., Cerqueira G. C., Mashiyama S. T., Al-Lazikani B., Andrade L. F., Ashton P. D., Aslett M. A., Bartholomeu D. C., Blandin G., Caffrey C. R., Coghlan A., Coulson R., Day T. A., Delcher A., DeMarco R., Djikeng A., Eyre T., Gamble J. A., Ghedin E., Gu Y., Hertz-Fowler C., Hirai H., Hirai Y., Houston R., Ivens A., Johnston D. A., Lacerda D., Macedo C. D., McVeigh P., Ning Z., Oliveira G., Overington J. P., Parkhill J., Pertea M., Pierce R. J., Protasio A. V., Quail M. A., Rajandream M. A., Rogers J., Sajid M., Salzberg S. L., Stanke M., Tivey A. R., White O., Williams D. L., Wortman J., Wu W., Zamanian M., Zerlotini A., Fraser-Liggett C. M., Barrell B. G., El-Sayed N. M. (2009) Nature 460, 352–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brunton V. G., MacPherson I. R., Frame M. C. (2004) Biochim. Biophys. Acta 1692, 121–144 [DOI] [PubMed] [Google Scholar]
- 39. Nagar B., Bornmann W. G., Pellicena P., Schindler T., Veach D. R., Miller W. T., Clarkson B., Kuriyan J. (2002) Cancer Res. 62, 4236–4243 [PubMed] [Google Scholar]
- 40. Shan Y., Seeliger M. A., Eastwood M. P., Frank F., Xu H., Jensen M. Ø., Dror R. O., Kuriyan J., Shaw D. E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li Q., Shen L., Xin T., Xiang W., Chen W., Gao Y., Zhu M., Yu L., Li M. (2009) BMC Dev. Biol. 9, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamanaka T., Ohno S. (2008) Front. Biosci. 13, 6693–6707 [DOI] [PubMed] [Google Scholar]
- 43. Newman R. A., Prehoda K. E. (2009) J. Biol. Chem. 284, 12924–12932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fichelson P., Huynh J. R. (2007) Prog. Mol. Subcell. Biol. 45, 97–120 [DOI] [PubMed] [Google Scholar]
- 45. Baumgartner M., Weiss A., Fritzius T., Heinrich J., Moelling K. (2009) Exp. Cell Res. 315, 2888–2898 [DOI] [PubMed] [Google Scholar]
- 46. Singh P. K., Hollingsworth M. A. (2006) Trends Cell Biol. 16, 467–476 [DOI] [PubMed] [Google Scholar]
- 47. Bafna S., Kaur S., Batra S. K. (2010) Oncogene 29, 2893–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schroeder J. A., Thompson M. C., Gardner M. M., Gendler S. J. (2001) J. Biol. Chem. 276, 13057–13064 [DOI] [PubMed] [Google Scholar]
- 49. Matsunami H., Buck L. B. (1997) Cell 90, 775–784 [DOI] [PubMed] [Google Scholar]
- 50. Krautz-Peterson G., Bhardwaj R., Faghiri Z., Tararam C. A., Skelly P. J. (2010) Parasitology 137, 485–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dissous C., Grevelding C. G. (2011) Trends Parasitol. 27, 59–66 [DOI] [PubMed] [Google Scholar]
- 52. Doenhoff M. J., Cioli D., Utzinger J. (2008) Curr. Opin. Infect. Dis. 21, 659–667 [DOI] [PubMed] [Google Scholar]
- 53. Stothard J. R., Chitsulo L., Kristensen T. K., Utzinger J. (2009) Parasitology 136, 1665–1675 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.