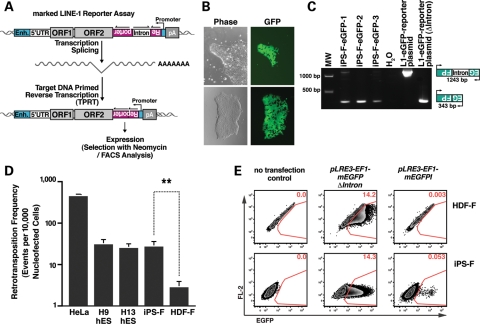
Figure 4.
The retrotransposition efficiency of an engineered human L1 is higher in pluripotent stem cells than in adult HDFs. (A) An overview of the L1 retrotransposition assay. An RC-L1 is tagged with a retrotransposition indicator cassette (mneoI or mEGFPI). The indicator cassette contains an antisense copy of a selectable (NEO) or detectable (EGFP) reporter gene that is disrupted by an intron in the same transcriptional orientation as the L1 (13,52). The reporter gene contains a heterologous promoter (SV40 for the mneoI and UBC for the mEGFPI) and a poly (A) signal. An EF1a or UBC promoter (Enh, enhancer) drives L1 expression. The reporter gene can only be activated when L1 RNA is reverse transcribed, integrates into genomic DNA and is expressed from its own promoter (13). (B) An undifferentiated iPS-F colony harboring a L1 retrotransposition event from the pLRE3-EF1-mEGFPI reporter. (C) L1 retrotransposition events are stably integrated into the genome. Genomic DNA isolated from EGFP-positive colonies after nucleofection served as a PCR template. The 343-bp PCR product indicates the spliced tagged L1 (insertion); the 1243-bp product contains the intron (vector). (D) L1 retrotransposition efficiencies in different cell lines. Cells were transfected with pLRE3-EF1-mEGFPI reporter or the pLRE3-EF1-mEGFP(▵intron) plasmid as a control. Cells nucleofected with pLRE3-EF1-mEGFPI were harvested 4 days after nucleofection; cells nucleofected with pLRE3-EF1-mEGFP(▵intron) were harvested 2 days after nucleofection at the peak of EGFP expression. The frequency of new integrations (EGFP-positive cells) was analyzed by flow cytometry. For hESCs or iPSCs, ∼1–2 × 106 cells/sample were analyzed; for HDF cells, ∼0.25–0.5 × 106 cells/sample were assayed. The retrotransposition frequency was calculated as the number of EGFP-positive cells after nucleofection with the pLRE3-EF1-mEGFPI construct divided by the number of EGFP-positive cells after nucleofection with the pLRE3-EF1-mEGFP(▵intron) control plasmid. The experiments were repeated at least three times. Values are the mean ± SEM. **P≤ 0.01. (E) Flow cytometry plots of iPS-F iPSCs and HDF-F cells either untransfected or transfected with pLRE3-EF1-mEGFPI reporter (harvested 4 days post-transfection) or the pLRE3-EF1-mEGFP(▵intron) control plasmid (harvested 2 days post-transfection at the peak of EGFP expression). For iPSCs, ∼1 × 106 cells/sample were analyzed; for HDFs, ∼0.5 × 106 cells/sample were assayed. Cells were plotted against an empty channel (FL-2) to eliminate background autofluorescent cells and to increase sensitivity.