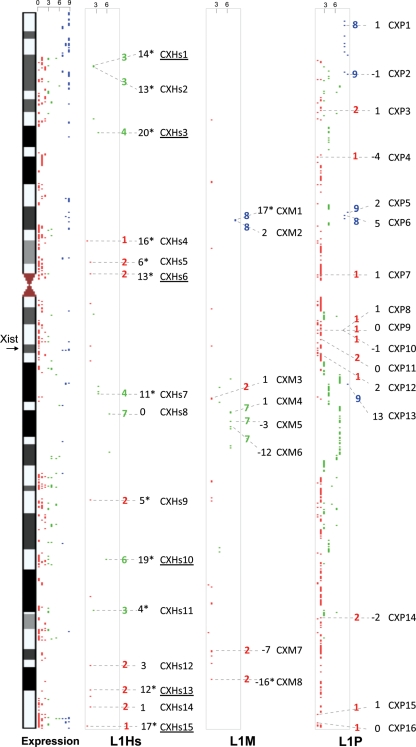
Figure 2.
Relative positions of the full-length (>6 kb) L1Hs, L1P and L1M on the X chromosome. The studied regions in this study are labeled with dashed line. Expression data from Carrel and Willard (57) are presented in the first column, where scale of expression is 0–9 (number of cell lines with inactive X that were expressing a given gene); we empirically considered score of 0–2 to be inactivated (red), 3–6 to be middle inactivated (green) and 8–9 escaping inactivation (blue). In the second, third and forth columns are the L1Hs, L1M and L1P full-length copies (>6 kb) represented by dots. An inactivation score for every locus was empirically calculated according to the position of the LINE-1 sequence relative to the studied transcripts represented in column 1, thus a 1 score is given for a locus in a gene that is inactivated, 2: between two inactivated genes, 3: between one inactivated gene and one middle inactivated gene, 4: between one inactivated gene and one non-inactivated gene, 5: between two middle inactivated genes, 6: in a middle inactivated gene, 7: between one middle inactivated gene and non-inactivated gene, 8: between two non-inactivated gene, 9: in a gene that is non-inactivated. The inactivation score is given right to the given studied L1 dot, followed by the male–female average methylation difference, followed by the name of the loci. The statistically significant difference between the males and females is labeled with a star. The underlined loci represent the ones studied in all four groups of samples.