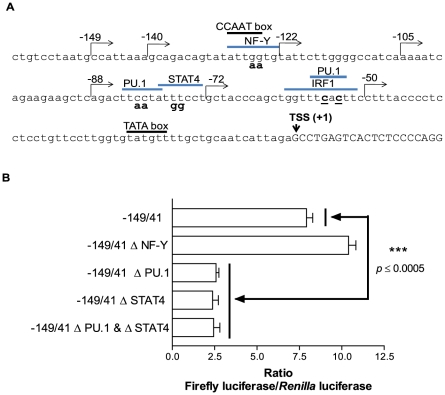
Figure 3. Location of putative transcription factor binding sites on FPR1 promoter.
A. Sequence analysis using PROMO3 software identified certain transcription factors commonly expressed in myeloid cells as putative regulators of FPR1 transcription. The numbers indicate the first nucleotide of the various promoter constructs in relation to the transcriptional start site (TSS). The −56 and −54 SNPs are underlined and the various mutations of putative transcription factor binding sites are in bold. B. Site-directed mutagenesis of the putative PU.1 and STAT4 binding sites resulted in a significant decrease in firefly luciferase activity. U937 cells were co-transfected with the indicated wild-type and promoter mutant constructs and pRL-TK to normalize for transfection efficiency. Data show the mean ratios from three experiments ± S.E.M. One-way analysis of variance showed that differences in luciferase activity among the constructs were significant (p value<0.0001), and unpaired t test showed that the luciferase activities of each of the PU.1 and STAT4 mutant constructs were significantly lower than that of the wild-type construct, p value≤0.0005.