Figure 6.
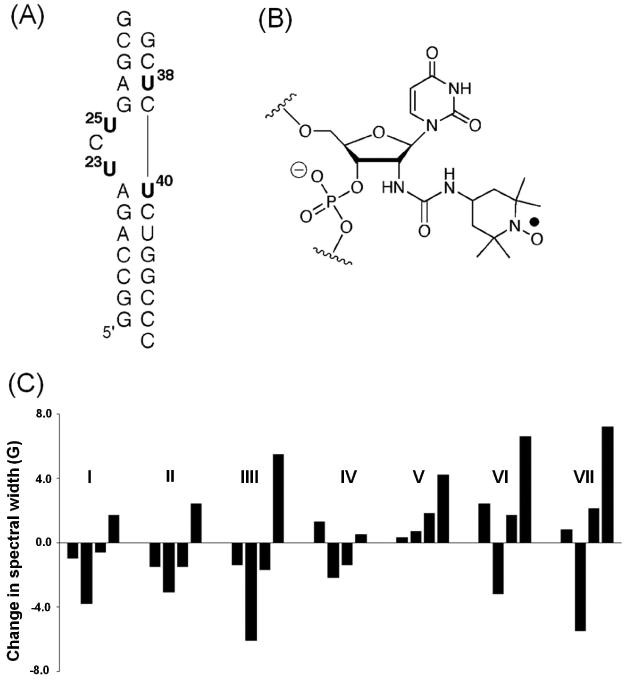
SDSL studies of the TAR RNA. (A) The secondary structure of the TAR RNA construct used in the study, with the four labeled uridines shown in bold. (B) Chemical structure of the nitroxide-labeled uridine. (C) An example of variations of dynamic signatures, which are represented by differences in the effective hyperfine splitting obtained from measured X-band cw-EPR spectra, upon TAR binding to various small molecule ligands (I: Hoechst; II: DAPI; III: berenil; IV: CGP40336A; V: neomycin; VI: guanidine neomycin; VII: arginiamide). Figure adopted from Ref. 19 with permission.
