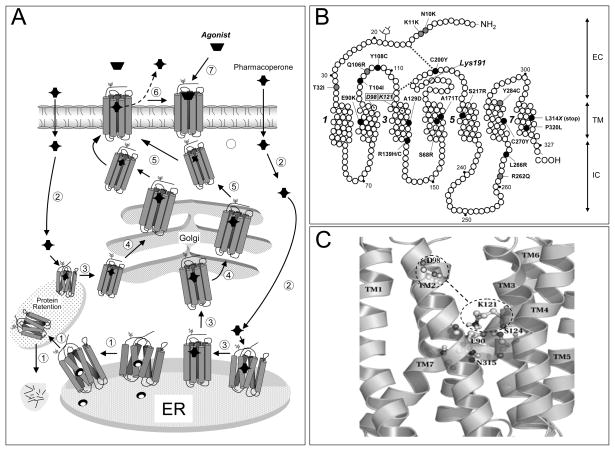
Fig. 1.
A: Quality control system in the endoplasmic reticulum (ER). Newly synthesized proteins (e.g. GPCRs) are translocated to the lumen of the ER where folding is facilitated by molecular chaperones (oval structures); if folding fails, misfolded proteins are retained in the ER and targeted for degradation by cytosolic proteasomes that cleaved the protein into fragments (short lines) (step1). Pharmacoperones diffuse into the cell (step 2) and selectively bind the misfolded protein to influence folding (step 3) and allow correct routing to the Golgi complex for further processing (e.g. glycosylation) (step 4). Previously synthesized misfolded proteins, retained by the QCS, may still be rescued by pharmacoperones (steps 2 and 3 at left). Mature proteins are then delivered to the cell surface membrane (step 5), where the pharmacoperone is dissociated from the receptor protein (step 6) allowing the receptor to interact with its cognate ligand (step 7). B: Location of the inactivating mutations in the human GnRHR. Grey ovals are mutations that lead to partial hypogonadotropic hypogonadism, whereas black ovals are mutations that provoke complete hypogonadotropic hypogonadism. Also shown is the location of K191 at the second extracellular loop, which destabilizes the formation of the C14-C200 bridge, as well as the location of D98 (in TM2 2) and K121 (in TM3) that form the surrogate D98-K121 salt bridge upon pharmacoperone action. Transmembrane domains 1, 3, 5, and 7 are numbered. EC: Extracellular domain; TM: Transmembrane domains; IC: Intracellular domains. C: Enlarged diagram showing specific interactions between E90 (in TM2), N315 (in TM7), K121 (in TM3) and S124 (in TM3), which form a microdomain important for GnRHR stability. D98 and K121 are shown within the dashed line ovals; the dashed line between the ovals represents the surrogate salt bridge that is formed upon pharmacoperone binding.