Fig. 1.
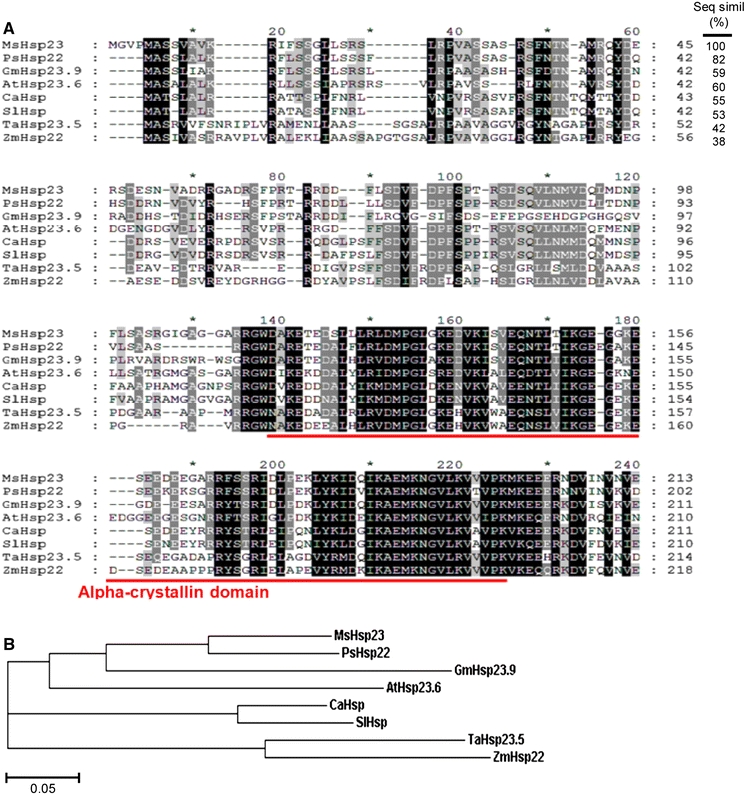
Multiple sequence alignment and phylogenetic analysis of MsHSP23. a Alignment of MsHSP23 with msHSPs homologs from pea (PsHSP22, P46254), soybean (GmHSP23.9, AAB03096), Arabidopsis (AtHSP23.6, NP_194250), chilli pepper (CaHSP, ADJ57588), tomato (SlHSP, BAA32547), wheat (TaHSP23.5, AAD03604) and corn (ZmHSP22, AAV32521) using the ClustalW and GeneDoc programs. The numbers at the right indicate the amino acid residue position. Fully identical residues are shaded in black, six identities in gray, and five identities in light gray. The conserved alpha-crystallin domain is indicated by a line. b The phylogenetic relationships between MsHSP23 and other msHSPs homologs. The phylogenetic tree shows a graphical representation of the evolutionary relationships and was constructed using the EBI-ClustealW2 algorithm
