Table 1.
Structures of fragments that bind solely H4R (1–3), solely 5-HT3AR (4–6) and both H4R and 5-HT3AR (7–11)
| # | H4R/5-HT3AR | Structure | Affinity (pKi) |
|
|---|---|---|---|---|
| H4Ra | 5-HT3ARc | |||
| 1 |  |
7.0 ± 0.1 | n.a. | |
| 2 |  |
6.2 ± 0.1 | n.a. | |
| 3 |  |
6.7 ± 0.0 | n.a. | |
| 4 |  |
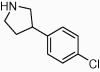 |
n.a. | 6.1 ± 0.2 |
| 5 |  |
n.a. | 6.0 ± 0.0 | |
| 6 |  |
n.a. | 6.1 ± 0.1 | |
| 7 |  |
6.2 ± 0.0 | 6.6 ± 0.3 | |
| 8 |  |
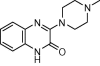 |
7.2 ± 0.0 | 7.9 ± 0.3 |
| 9 |  |
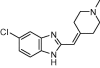 |
6.1 ± 0.1b | 8.8 ± 0.1 |
| 10 |  |
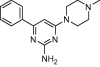 |
8.2 ± 0.1 | 5.9 ± 0.1 |
| 11 |  |
6.2 ± 0.1b | 5.9 ± 0.3 | |
n.a.: Non active.
Measured by displacement of [3H]histamine binding using membranes of HEK293 cells transiently expressing the human H4R. pKi’s are calculated from at least three independent measurements as the mean ± SEM.
Determined using membranes of SK-N-MC cells transiently expressing the human H4R. pKi’s are calculated from at least three independent measurements as the mean ± SEM.
pKi: Measured by displacement of [3H]granisetron binding using membranes of HEK293 cells expressing the human 5-HT3AR. pKi’s are calculated from at least two independent measurements as the mean ± SEM.
