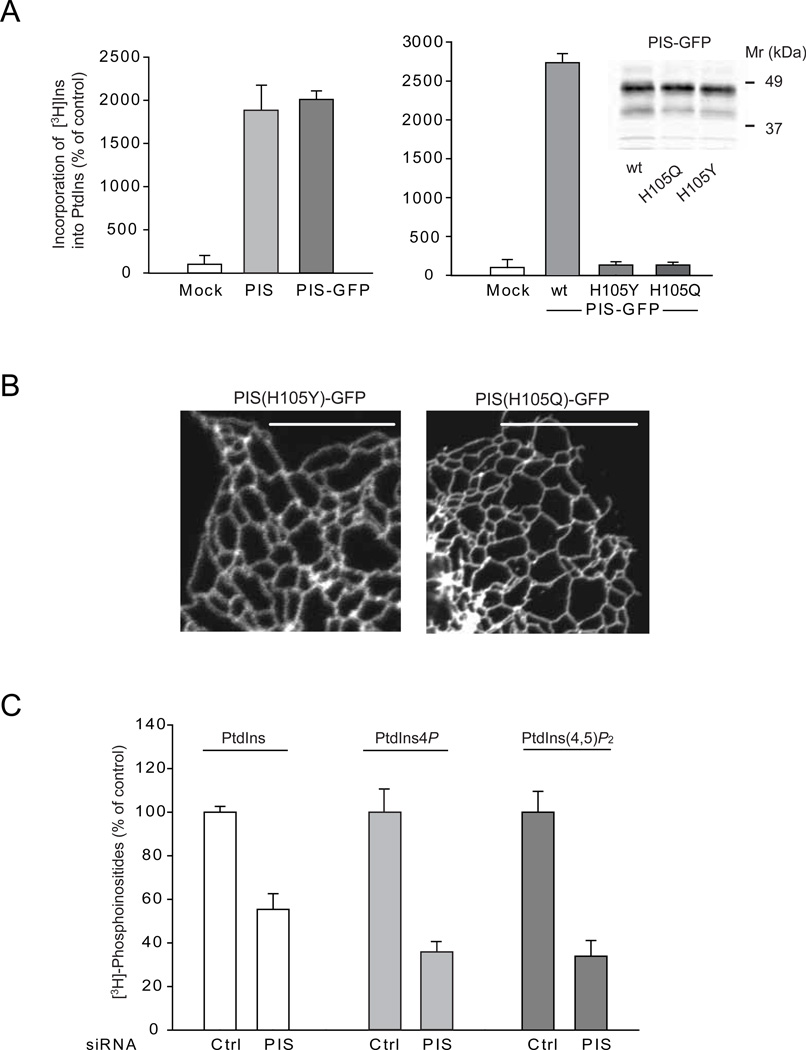
Figure 3. Relationship between PIS activity and localization and effects of PIS knock-down on cellular phosphoinositide levels.
(A) Crude membranes prepared from COS-7 cells expressing either an empty vector, the untagged PIS or PIS-GFP or its mutants (H105Y and H105Q) were assayed for PtdIns synthase activity as described under Methods. The incorporation of myo-[3H]inositol into PtdIns using CDP-DAG as a substrate was significantly increased in cells transfected with either forms of wild-type PIS but not the mutant enzymes. The results of a representative experiments are shown performed in duplicates. The inset on the right shows equal expression of the wild-type and mutant PIS-GFP proteins. (B) Cellular localization of mutant PIS-GFP enzymes (H105Y and H105Q) expressed in COS-7 cells. Note the lack of the mobile PIS positive structures. (C) HEK293-AT1 cells were treated with control siRNA or PIS siRNA for 3 days and labeled with myo-[3H]inositol for 24 hrs as described under Methods. Labeled lipids were extracted from the cell pellets, separated by TLC, and analyzed by a densitometry of exposed films. Error bars indicate SEM (from 3 independent experiments, each performed in duplicates). In other experiments where the separated lipids were eluted and counted with a scintillation counter, the labeling of PtdIns4P and PtdIns(4,5)P2 were 6 and 4 %, respectively, of that of PtdIns.