Figure 7.
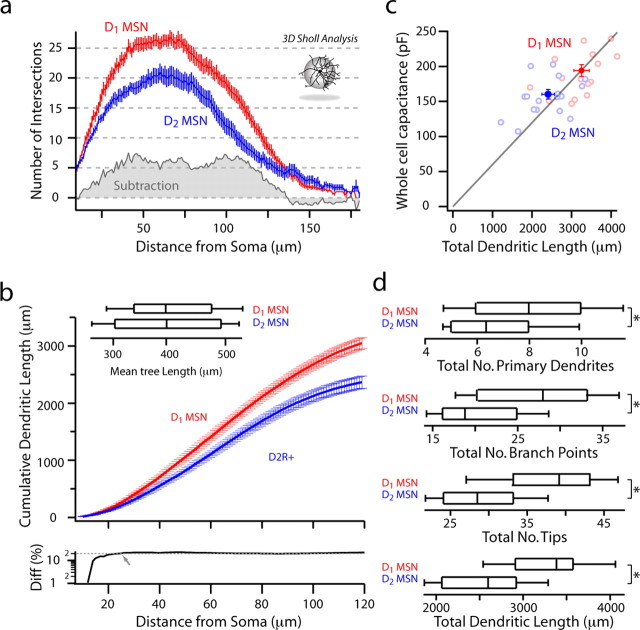
Analysis of anatomical differences between reconstructed D1 and D2 MSNs. a, A three-dimensional Sholl analysis of biocytin-filled and reconstructed neurons from P35–P45 BAC transgenic mice. Data are shown as mean (±SEM) number of intersections at 1 μm eccentricities from the soma for 15 D1 and 16 D2 MSNs. D1 MSNs have a more highly branched dendritic tree, as indicated by the increased number of intersections and positive subtracted area (gray shading). b, Mean cumulative dendritic length at 1 μm eccentricities for D1 and D2 MSN populations. Bottom, Note that the percentage difference (Diff) between populations in cumulative total dendritic length increases and remains at ∼20% (arrow and fit line) within 30 μm from the soma. Inset, The total dendritic length in each cell/number of primary dendrites is not significantly different between populations (D1 MSN: median, 398.8 μm, n = 15; D2 MSN: median, 400.5 μm, n = 16). c, Whole-cell capacitance is positively correlated to total dendritic length (rS = 0.45, p < 0.05). d, D1 MSNs have significantly more primary dendrites (D1 MSN: median, 8, n = 15; D2 MSN: median, 6, n = 16; p < 0.05), branch points (D1 MSN: median, 28, n = 15; D2 MSN: median, 19, n = 16; p < 0.05), tips (D1 MSN: median, 38, n = 16; D2 MSN: median, 28, n = 15; p < 0.001), and total dendritic length (D1 MSN: median, 3385.6 μm, n = 15; D2 MSN: median, 2878.3 μm, n = 16; p < 0.001). *Statistical significance.