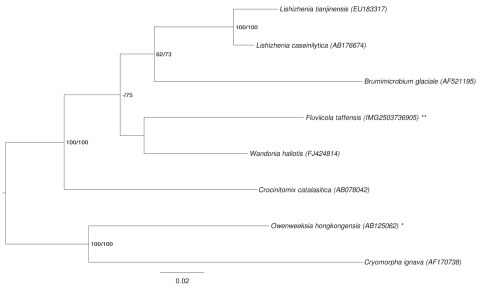
Figure 1.
Phylogenetic tree highlighting the position of F. taffensis relative to the type strains of the other species within the family Cryomorphaceae. The tree was inferred from 1,429 aligned characters [13,14] of the 16S rRNA gene sequence under the maximum likelihood (ML) criterion [15]. Rooting was done initially using the midpoint method [16] and then checked for its agreement with the current classification (Table 1). The branches are scaled in terms of the expected number of substitutions per site. Numbers adjacent to the branches are support values from 300 ML bootstrap replicates [17] (left) and from 1,000 maximum parsimony bootstrap replicates [18] (right) if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [19] are labeled with one asterisk, those also listed as 'Complete and Published' (as well as the target genome) with two asterisks.