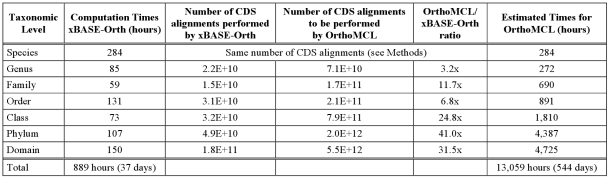
Figure 3. Computation times and estimated speedup compared to OrthoMCL.
By adopting practical accuracy/speed trade-off xBASE-Orth allows for acceptable computational time using reasonable hardware resources and is predicted to be about 15 times faster compared to direct application of OrthoMCL. The vast majority of the computation time is spent performing CDS alignments and operating on the smaller pan-genomes (xBASE-Orth) rather than on the full CDS collection (OrthoMCL) results in significant time advantage. It is more prominent at higher taxonomic levels, where the difference between pan-genome and full CDS collection sizes increases.