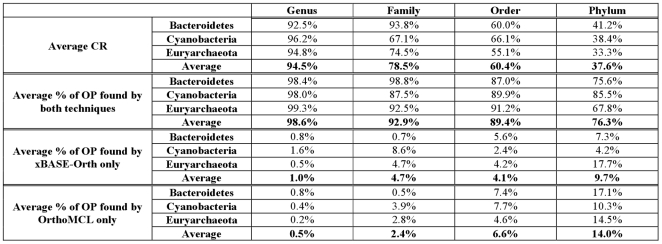
Figure 4. xBASE-Orth vs. OrthoMCL comparison.
xBASE-Orth has a significant speed advantage over direct application of OrthoMCL which comes at the possible cost of decreased accuracy. We compared the performance of the two approaches over three distinct phyla – Bacteroidetes (40 complete genomes), Cyanobacteria (41 genomes) and Euryarchaeota (62 genomes), computing orthologs at genus, family, order and phylum level. At higher taxonomic levels the pan-genome sizes are significantly smaller compared to the full CDS collections - about half at order level and only about one third at phylum level for the datasets analyzed here (CR stands for Compression Ratio = [Number of CDSs in pan-genomes used by xBASE-Orth/Number of CDSs used by OrthoMCL] * 100%). Hence, at higher levels each CDS in a pan-genome is a representative of a larger set of orthologous/paralogous CDSs, plausibly becoming less sensitive and specific. Compared to the OrthoMCL results, it appears that on average the xBASE-Orth results contain from 1% (at genus level) to 9.7% (phylum) additional ortholog pairs, while failing to detect from 0.5% (genus) to 14% (phylum) OrthoMCL pairs.