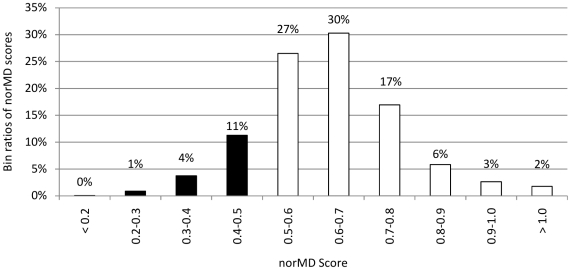
Figure 5. Distribution of norMD scores for the 2,384 multiple alignments.
To investigate the specificity of xBASE-Orth we selected 2,384 CDSs such that: i) each CDS has at least one ortholog at each taxonomic level; ii) it has no more than 150 orthologs in total; and iii) the ortholog groups (OGs) for each of the 2,384 CDSs are disjoint. For each of the CDSs, we fetched the orthologs predicted by xBASE-Orth, performed multiple alignment of the sequences with ClustalW2, and scored the alignment with the norMD tool. The x-axis lists the chosen bin ranges for the norMD value. The y-axis depicts the distribution of the observed norMD values across the bins. A value of 0.5 or greater is considered to be the cut-off value for a good multiple alignment, indicating high level of sequence similarity. A vast majority (84%) of the chosen 2,384 OGs exhibit suitable sequence similarity and align well, producing norMD values ≥0.5. It is worth noting that on average about 60% of the orthologs fetched for each of the 2,384 CDSs are at phylum and domain level – i.e. xBASE-Orth exhibits good specificity even at large evolutionary distances.