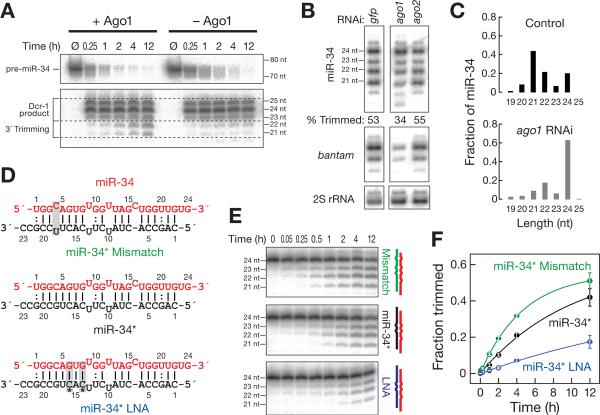
Figure 2. Trimming of miR-34 requires Ago1 and is limited by the removal of the miRNA* strand.
(A) 52 32P-radiolabeled pre-miR-34 was incubated in 0–2 h embryo lysate or lysate immunodepleted of Ago1. Products were resolved by denaturing polyacrylamide gel electrophoresis. (B) DsRNA-triggered RNAi targeting Ago1, but not Ago2, decreased trimming of miR-34, compared to treatment with a control dsRNA targeting GFP. “Trimmed” indicates the fraction of all miR-34 corresponding to 21 and 22 nt isoforms. The bantam miRNA and 2S rRNA served as controls. (C) The fraction of long miR-34 isoforms, measured by high throughput sequencing, increased when S2 cells were depleted of Ago1 by RNAi. Only isoforms with the annotated miR-34 52 end were analyzed. The abundance of miR-34 in the two libraries was 3499 ppm (control) and 4506 ppm (ago1 RNAi). (D, E) Synthetic duplexes of 52 32P-radiolabeled miR-34 (red) paired to variants of miR-34* (D) were incubated in 0–2 h embryo lysate (E), and the products analyzed by denaturing polyacrylamide gel electrophoresis. (F) Mean ± standard deviation for three independent replicates of the experiment in (E). See also Figure S2.