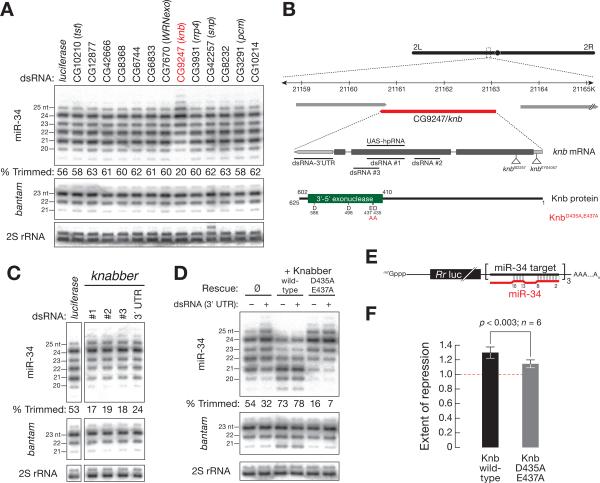
Figure 3. The 32-to-52 exoribonuclease Knabber (CG9247) trims miR-34, enhancing miR-34 function in S2 cells.
(A) S2 cells were transfected with dsRNA against a panel of predicted exonucleases and the effect on miR-34 length analyzed by high resolution Northern hybridization. bantam and 2S rRNA served as controls. The fraction of miR-34 trimmed to 21–22 nt is indicated below each lane. (B) The predicted structure of the knabber (CG9247) gene, mRNA, and protein. (C) S2 cells were transfected with three dsRNAs targeting the second exon or the 32 UTR of knabber as indicated in (B). All four dsRNAs decreased miR-34 trimming, relative to a control dsRNA targeting firefly luciferase. bantam and 2S rRNA served as controls. (D) S2 cells stably expressing wild -type or D435A,E437A mutant Knabber were transfected with dsRNA targeting the 32 UTR of endogenous knabber, and the effect on miR-34 trimming measured. bantam and 2S rRNA served as controls. (E) Reporter construct used in (F). The three miR-34 binding sites pair with miR-34 nucleotides 2–8 and 13–15, mimicking typical animal miRNA binding sites [54]. Rr luc, Renilla reniformis luciferase. (F) Knabber trimming of miR-34 enhances miRNA function. Repression by miR-34 in S2 cells expressing wild-type or D435A,E437A mutant Knabber was measured by blocking miR-34 using a 22-O-methyl-modified anti-miRNA oligonucleotide and measuring the increase in Rr luciferase expression compared to a control oligonucleotide targeting let-7, a miRNA not normally expressed in S2 cells. See also Figure S3.