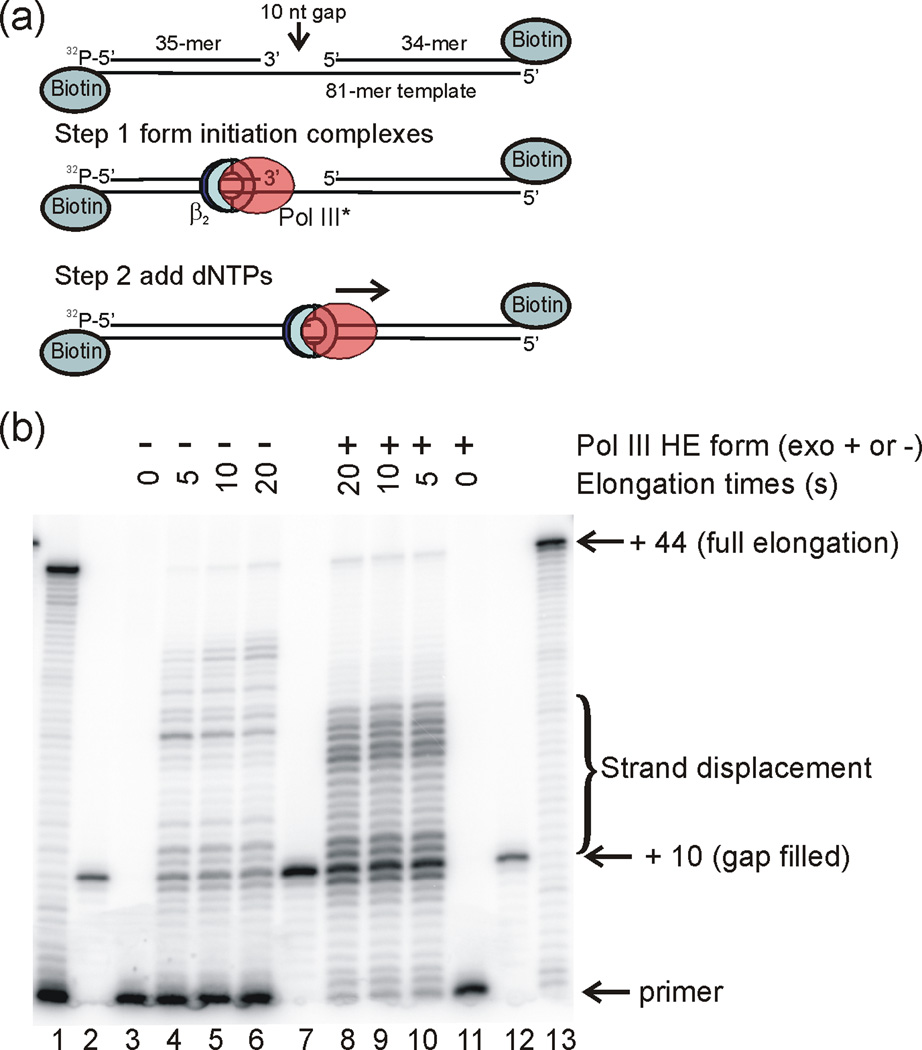
Fig. 3.
Pol III HE rapidly fills a 10 nt gap and partially displaces a blocking oligonucleotide. (a) 32P-5′ end labeled primer 35-mer was hybridized to a template 81-mer with a 10 nt gap in front of blocking oligonucleotide 34-mer. This primer-template was incubated with Pol III HE and ATP for 60 s to form initiation complexes, followed by addition of dNTPs to initiate primer elongation for the 32P-labeled complexes. (b) Denaturing polyacrylamide gel analysis of the rate of nucleotide incorporation. The gel lanes were loaded with 1) unblocked primer-template fully extended by Pol III HE; 2) 32P-5′ end labeled 45-mer corresponding to a fully gap-filled product, annealed to the 81-mer template; 3) unextended 10 nt gap primer-template; 4) 10 nt gap primer-template after 5 s extension time; 5) 10 nt gap primer-template after 10 s extension time; 6) 10 nt gap primer-template after 20 s extension time; 7) 45-mer annealed to the 81-mer template (4-fold more sample than loaded than in lanes 2 and 12); 8) 10 nt gap primer-template after 20 s extension time; 9) 10 nt gap primer-template after 10 s extension time; 10) 10 nt gap primer-template after 5 s extension time; 11) unextended 10 nt gap primer-template; 12) 45-mer annealed to the 81-mer template; 13) unblocked primer-template fully extended by Pol III HE. The samples in lanes 1 and 4 – 6 were extended by exonuclease-deficient Pol III HE. The samples in Lanes 8 – 10 and 13 were extended by wild-type Pol III HE.