Abstract
Intracellular pathogens such as Listeria monocytogenes and Rickettsia rickettsii move within a host cell by polymerizing a comet-tail of actin fibers that ultimately pushes the cell forward. This dense network of cross-linked actin polymers typically exhibits a striking curvature that causes bacteria to move in gently looping paths. Theoretically, tail curvature has been linked to details of motility by considering force and torque balances from a finite number of polymerizing filaments. Here we track beads coated with a prokaryotic activator of actin polymerization in three dimensions to directly quantify the curvature and torsion of bead motility paths. We find that bead paths are more likely to have low rather than high curvature at any given time. Furthermore, path curvature changes very slowly in time, with an autocorrelation decay time of 200 s. Paths with a small radius of curvature, therefore, remain so for an extended period resulting in loops when confined to two dimensions. When allowed to explore a three-dimensional (3D) space, path loops are less evident. Finally, we quantify the torsion in the bead paths and show that beads do not exhibit a significant left- or right-handed bias to their motion in 3D. These results suggest that paths of actin-propelled objects may be attributed to slow changes in curvature, possibly associated with filament debranching, rather than a fixed torque.
Introduction
The polymerization of cross-linked, dendritic networks of actin filaments is an important biological phenomenon that drives such diverse processes as cell crawling, phagocytosis and morphogenesis [1–3]. These complex networks are organized by a number of proteins, including nucleation-promoting factors that trigger the formation of nascent filaments off the side of existing network filaments [1, 2, 4]. Bacteria such as Listeria monocytogenes and Rickettsia rickettsii propel themselves during infection by expressing a single protein on their cell surfaces, ActA and RickA, respectively. Both of these factors stimulate actin growth by activating the branching complex Arp2/3, which forms the junction of the characteristic 70° branches within the actin network [5]. Actin polymerization at the cell’s rear gives rise to a characteristic comet tail that propels the cell through the host cytoplasm. Coating small spheres with either ActA or RickA gives rise to actin propulsion in eukaryotic cellular extracts with an indistinguishable architecture of Y-branch crosslinking of actin filaments [5, 6]. These networks appear similar to those found in motile eukaryotic cells [7]. As a result, moving beads have become a model system for studying actin-based motility in general. Interestingly, viable Rickettsia cells are driven by comet tails comprised of straight filaments that lack both branching and the incorporation of the Arp2/3 proteins [8]. Presumably, these cells express other factors that modify the branched network close to the bacterial surface and produce a straight-filament tail.
Several biophysical models have been developed to describe the connection between network growth and the generation of force [9–12]. Despite the extensive theoretical and experimental investigation of polymerization forces and velocities [13, 14], less is understood about the changes in direction of network growth.
Comet tails are often highly curved and can propel cells and beads in nearly circular or looping paths [12, 15, 16]. Phenomenological description of the shapes of the comet tails and the trajectories of actin-propelled objects has been used to gauge the effect of biophysical and biochemical perturbations on the process of network formation and maintenance [17–19]. In recent work that examined moving L. monocytogenes cells confined to two dimensions, Shenoy et al postulate that a roughly constant torque drives cells in looping paths [20]. A detailed quantification of actin-propelled trajectories in three dimensions can yield additional information about all the forces and torques that, on average, act on the moving object. Indeed, theoretical treatment of network curvature has been used to place limits on the number and placement of pushing filaments at a load surface [21]. However, the lack of three-dimensional data on this subject has hindered progress in understanding the mechanisms controlling curvature.
Traditional measurements of object trajectories face two main difficulties. First, experiments often track the in-plane object movements or fluorescence of actin in the network, inherently two-dimensional (2D) techniques that do not allow one to monitor the full 3D complexity of the trajectories. Second, due to the limited depth of field of conventional light microscopy, a micron-sized gap between a coverslip and microscope slide is used to restrict out-of-plane motion, leading to pseudo-2D observations [5, 16, 17, 22]. Motivated by these limitations and an interest in the mechanism governing movement direction, we sought to measure the 3D trajectories explored by actin-propelled beads in large chambers using three-dimensional laser tracking.
Methods
Motility assays
Experiments were performed as described previously [23]. Briefly, carboxylated polystyrene beads 792 + 23 nm in diameter were coated with the nucleation factor RickA purified from R. rickettsii (a gift from Matthew Welch, University of California, Berkeley). The assay mixture, containing beads in Xenopus laevis egg cytoplasmic extract supplemented with an ATP-regenerating mix and Rhodamine-labeled actin, was added to a microscope slide immediately after preparation. When an active bead with an attached fluroescently-labeled comet tail was found, the bead was moved to the center of the laser focus and feedback was started. Each bead was monitored until motion ceased (usually caused by the tail suddenly detaching from the bead) up to a maximum of 30 min. A new slide was prepared for each single bead measurement.
We used chambers of two thicknesses to monitor bead motion in pseudo-2D and -3D environments. Approximately 2 μm chambers were created using 2.1 + 0.1 μm polystyrene beads as spacers between a glass slide and coverslip. 80 μm thick chambers were constructed by separating a microscope slide and coverslip with double-sided tape.
Beads moved with a slightly slower velocity, υ, in the 2D environment than in 3D (table 1), most likely due to a viscous interaction with the top and bottom glass surfaces that affect the Brownian Ratchet mechanism of actin-based motility [23]. We found that bead velocities and the percentage of beads that successfully broke symmetry and formed comet tails varied between cellular extracts prepared at different times. As a result, all of the data presented here was measured using material from a single cytoplasmic extract preparation which was aliquoted and frozen. In the thicker chambers, the average velocity from each bead ranged from 58 nm s−1 to 90 nm s−1. The standard deviation in velocity for individual beads was ~14 nm s−1. However, velocity fluctuations appeared random, beads did not systematically speed up or slow down over course of the experiment. In 2D, the velocity was heavily affected by the position of the bead along the z-direction [23].
Table 1.
Motility parameters for different geometries.
| L (μm) | υ (nm s−1)* | κRMS (μm−1)† | ζ (μm)† | τ (μm−1)† |
|---|---|---|---|---|
| 2 | 58 ± 38 | 0.22 ± 0.04 | 11 ± 6 | – |
| 80 | 76 ± 10 | 0.16 ± 0.03 | 16 ± 9 | −0.02 ± 0.03 |
±SD.
±SEM.
Three-dimensional laser tracking
Our instrument uses a single 809 nm wavelength diode laser to measure microsphere position using a position-sensitive detector that monitors forward scattered light in the back focal plane [24, 25]. We use a nano-positioning stage capable of moving the sample in three dimensions over hundreds of microns at a bandwidth of approximately 100 Hz. In addition, we record fluorescence images concurrent with optical tracking to confirm that bead motion corresponds to network growth (see figure 1 from [23]).
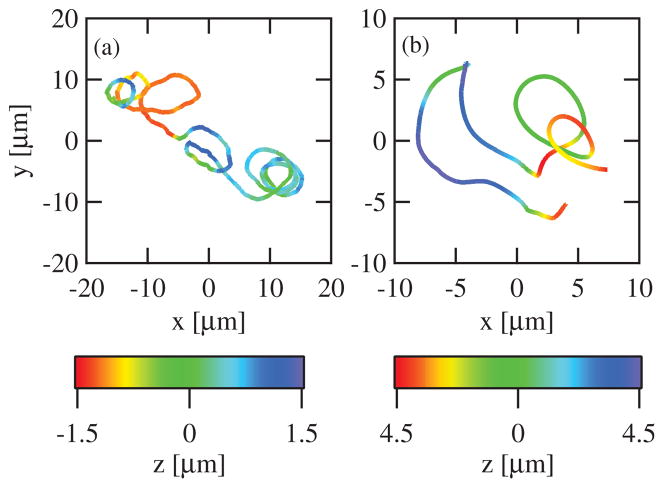
Figure 1.
Example trajectories of 3D bead motion. Data are shown for a bead moving in the 2 μm (a) and 80 μm (b) chambers. The color scale denotes the z-position, the position along the microscope optical axis.
During an experiment, a feedback routine, implemented in software, allows us to track actin-propelled beads over very long distances. 3D photodiode voltage signals [25–27] are sampled at 2 kHz and converted into 3D position in realtime using a fifth-order 3D polynomial [25]. The measured bead coordinates are then saved to disk and used to update the position of the microscope stage such that the microsphere remains in the center of the laser focus. By combining the stage position data from the capacitive sensor with the much faster bead position data from the laser tracking, we achieve approximately one nanometer resolution at a bandwidth of 1 kHz [23].
Very low laser powers were used to minimize external loads that might perturb actin network dynamics. These powers yielded typical forces of less than 0.01 fN.
Data analysis
Time derivatives of the x-, y- and z-positions versus time records were performed using a fourth-order Savitsky–Golay filter with a time constant of 1 s [28]. By trial and error, we found that a 1 s time constant gave reliable time derivatives of the position without amplifying noise for the higher orders. We confirmed that changing this time constant does not significantly alter the values reported here. Time derivatives were then used to calculate the curvature and torsion as defined by equations (1) and (2).
Results and discussion
We used a variant of 3D optical-force microscopy to track 0.8 μm diameter beads coated with the nucleation-promotion factor RickA moving in X. laevis egg cytoplasmic extract. Using 2 μm and 80 μm thick chambers, we created pseudo-2D and -3D environments to directly test the effect of the reduction of dimensionality. In the 2D chambers (n = 13, observation time ranged from 7.0 min to 17.6 min), beads proceeded in characteristically looping paths similar to previous observations (figure 1(a)) [15, 16]. Qualitatively, we observed cases of short-lived planar motion that resembled circles and figure-eights within the microscope plane. These paths were markedly different from those we observed using the 3D chambers (n = 15, observation time ranged from 9.9 min to 30.1 min), where planar motion was not observed (figure 1(b)). Instead, beads traveled in complex curved paths that, although often curved, rarely remained within the same plane, or traveled in a uniform helix for any substantial amount of time.
To analyze these paths in greater detail, we sought to quantify the shape of the measured bead trajectories mathematically. Up to an arbitrary translation and rotation, a geometric path in space can be fully described by two generalized curvatures which track the movement of the Frenet–Serret frame of the curve, termed the curvature and torsion. The curvature, κ, measures the deviance of a trajectory from a straight line, where 1/κ represents the radius of curvature at a given point. Using the coordinates of the trajectory with time, r(t), the curvature can be written as
| (1) |
The torsion, τ, measures the deviance of the trajectory from a planar curve and can be written as
| (2) |
If the torsion is zero, the trajectory lies completely in one plane. Conversely, a positive torsion corresponds to a right-handed helicity whereas a path with negative torsion has a left-handed helical component. For a helix of pitch, p, and radius, R, the magnitude of the torsion is given by
| (3) |
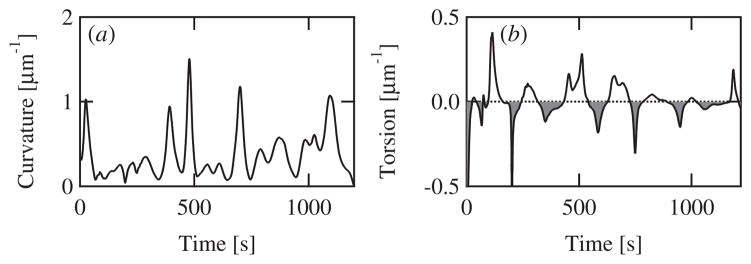
The instantaneous curvature and torsion over time calculated for the bead trajectory shown in figure 1(b) is displayed in figure 2.
Figure 2.
The calculated curvature (a) and torsion (b) as a function of time for the example trajectory shown in figure 1(b) from a bead moving in an 80 μm chamber. (b) Open shading denotes times during which the trajectory torsion is positive and thus moving with a right-handed component. Gray shading denotes left-handed sections. Note that the torsion crosses zero multiple times and thus the bead trajectory is not predominantly right or left handed.
We measured the curvature for beads moving in either pseudo-2D or -3D environments using chambers of two different thicknesses. The RMS curvature measured for these two situations, ~0.2 μm−1 (table 1), is within error of each other and is in good agreement with the curvature deduced from previous video-microscopy studies of the motion of both beads and bacteria of approximately 0.1 μm−1 [12, 16, 17].
To our knowledge, only one theoretical treatment of actin-driven motion has considered the molecular mechanism that produces looping trajectories. Rutenberg and Grant [21] showed that a small number of randomly distributed actin filaments pushing a load will give rise to an intrinsic curvature due to the nonzero instantaneous torque produced by summing the forces from each filament. For a uniformly-coated bead, this curvature is given by
| (4) |
where a is the bead radius and n is the number of pushing filaments. In this model, the distribution of curvatures, termed PI, depends only on the RMS value, and for a bulk, 3D geometry can be written as [21]
| (5) |
This distribution contains a single broad peak centered at (figure 3(a) dotted line), and thus beads are predicted to move with a ‘preferred’ curvature which leads to looping paths. Our 3D tracking experiments avoid the 2D projections usually found in light-microscopy measurements of moving beads, and thus it is appropriate for us to compare our data with the intrinsic PI distribution rather than the ensemble distribution, PE from [21].
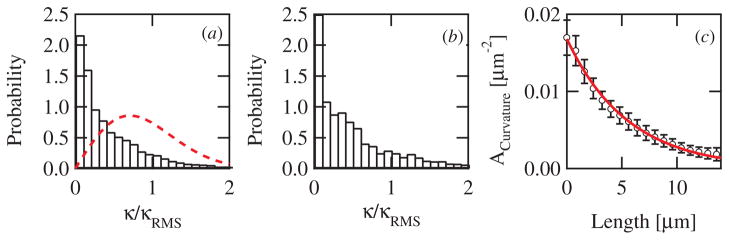
Figure 3.
(a) The 3D curvature probability distribution. The probability of measured curvatures for all beads using the 80 μm chambers is displayed (bars, n = 15). The curvature has been normalized by its RMS value from each run. For comparison, the distribution predicted by Rutenberg and Grant [21] is shown (red dotted line). (b) The normalized curvature distribution for all runs using the 2 μm chambers is displayed (n = 13). (c) Autocorrelation of the path curvature. The autocorrelation of the curvature as a function of path length for a single bead trajectory (open circles) and a fit of this data to a single exponential decay (red solid line).
To directly test this model we calculated the curvature probability distribution from our measured 3D trajectories in both sets of chambers (figures 3(a) and (b)). Rather than a peaked distribution, we find that both curvature distributions decay monotonically, with smaller curvatures, i.e. straighter path segments, more probable that curved ones. Notably, both the 80 μm and 2 μm thick chambers yielded qualitatively similar curvature distributions, implying that the mechanism of force generation and trajectory shape is most likely the same in each case. In contrast to the model of Rutenberg and Grant, our measured curvature distributions are not peaked at a nonzero curvature value yielding a poor match between model and experiment (figure 3(a)), and thus our data seem to rule out this mechanism of curvature formation.
Bead paths most often had low curvatures; 50% of the time the radius of curvature was greater than 25 μm. This feature is at least qualitatively reproduced by any model that includes a restoring force that opposes path curvature. One possibility is that a change in the actin density at the surface induces curvature, but that the response of growth to an asymmetric density rights the path. Such a feedback mechanism is similar in spirit to the autocatalytic mechanisms which having been employed previously to describe the affect of load on actin network growth [29].
If bead paths are most often straight, how can we reconcile the appearance of looping trajectories in the 2D chambers such as those shown in figure 1(a)? In the absence of a preferred curvature, loops might be created if network curvature can only change via slow dynamics. If the time to change path curvature is greater than the time to go around a loop, tloop = 2π/κυ, a path will necessarily complete a loop before significantly altering its trajectory. These dynamics may be associated, for instance, with the time required to significantly restructure the micro-architecture of the filaments.
We quantified the temporal dynamics of trajectory curvature by calculating the autocorrelation of the measured curvatures over time (figure 3(c)). The autocorrelation decays exponentially with an average decay constant, ζ, of 16 ± 9 μm for 3D movement, corresponding to an decay time of tdecay ~ 210 s. In the 2D chambers, the decay constant was lower than in 3D, measuring 11 ± 5 μm, but corresponding to approximately the same decay time, 190 s, because of the slower measured velocity.
From these data it is possible to describe why beads appear to move in looping paths when confined to 2D. While a path is most likely to be fairly straight at any instance in time, there is a small probability it will become highly curved. If a highly curved path is initiated, the slow curvature dynamics cause the path to remain highly curved for an extended period resulting in loops when confined to two dimensions. When allowed to explore a 3D space, where a constant curvature does not lead to planar loops, highly-curved path sections merely look ‘curly’ rather than looping.
What might cause the 200 s time scale, which is much slower than the characteristic times for actin monomer addition, ~10−2 s, or the addition of new filaments to the network, ~10−1 s [9]? While our beads are round and uniformly coated with RickA, there may be some residual asymmetry to the spatial distribution of the nucleator. Assuming that this density affects the shape and dynamics of network formation, a rotation of the bead relative to the network could yield a change in path curvature as discussed previously [21]. Therefore, it is possible that this time may correspond to the rotational diffusion time for a bead attached to a growing actin network. Similarly, if the distribution of active RickA molecules on the surface of the bead changes over time, e.g. by transient binding of an entire RickA molecule to the bead surface, population binding kinetics might govern the correlation time. Another explanation might involve architectural changes in the actin network itself. Recent evidence indicates that a growing actin network contains memory of past forces it has encountered [14]. The actin networks propelling the bead may actively remodel their micro-architecture in response to load and other factors on very slow time scales. Indeed, the half time for dissociation of Arp2/3 branched filaments from their mother filaments has been observed to be 500 s in the absence of ADF/cofilin and 30 s in the presence of ADF/cofilin [30]. We propose that the 200 s relaxation time we observe in the curvature autocorrelation function may represent the time-scale over which actin remodeling, perhaps due to network debranching, occurs.
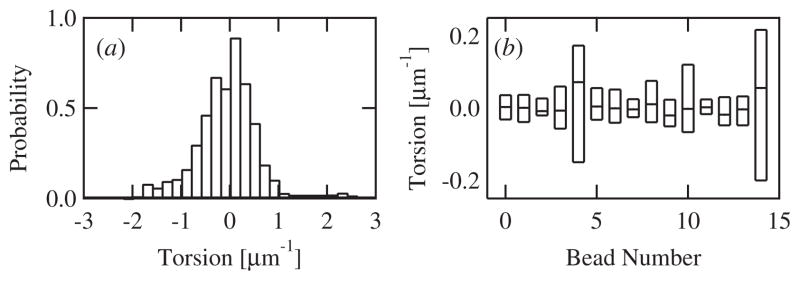
In addition to the curvature, we calculated the torsion for beads moving in 3D. The distribution of measured torsions from all beads analyzed together was roughly symmetric about zero with a small negative skew (figure 4(a)). The average torsion for all 3D runs was −0.02 ± 0.03, not significantly different than zero. While, brief sections of bead movement sometimes appeared helical (see, e.g., figure 1(b)), the torsion varied considerably, often crossing zero multiple times in a single run (figure 2(b)). We recovered similar results when individual beads were analyzed separately. None of the 15 beads we measured in the 3D chambers exhibited strictly positive or negative torsion over the course of the observation, each trajectory contained multiple right- and left-handed sections. Indeed, while the average torsion for individual beads ranged from −0.14 to +0.12, the distribution of torsions from each bead was wide including both positive and negative torsions (figure 4(b)). Overall our data show that beads do not move in consistently helical paths. In addition, we measured the autocorrelation of the torsion and found no significant correlation over time. The average autocorrelation decayed to zero with a time constant equal to the smoothing time constant used to calculate the derivatives of the position with time.
Figure 4.
(a) The torsion probability distribution. The torsion of measured curvatures for all beads using the 80 μm chambers is displayed (n = 15). (b) Box plot of the curvature distribution from each of the 15 individual beads whose data is compiled in (a). The three horizontal lines represent the 25th, 50th, and 75th percentiles of each set of torsions.
Our data are markedly different from the right-handed helical tails which have been observed with Listeria monocytogenes moving in high-speed brain extracts [31]. These cells move with a roughly constant torsion of ~0.15 μm−1 and are always right handed. These trajectories, along with data that moving Listeria cells rotate around their long-axis [32], suggest that actin-propelled beads, both the ones used in this study and ActA-coated beads from previous studies that do not move in helical paths [17], and Listeria might move by potentially different mechanisms. In particular, Listeria cells have been reported to produce a helical surface pattern of ActA [33], which may give rise to a network that applies a constant torque to moving cells, producing helical trajectories in 3D and the looping trajectories seen in 2D [20]. Uniformly coated bead from this and previous studies would not experience this torque.
Conclusion and outlook
We have presented experimental measurements of the 3D paths explored by actin-propelled beads. In a pseudo-2D environment, beads appear to move in looping paths as has been shown previously for measurements using video microscopy. In contrast, relaxing the restriction to 2D movement, we find that in 3D, beads do not move in looping paths, but still exhibit curved, complex structures. Differing from observations of moving Listeria cells, our beads do not move in helical paths in 3D environment, and instead we measure an average torsion of zero.
While these new data place constraints on the physical mechanisms that give rise to the movement of nucleator-coated beads, they also speak to the geometric persistence inherent in an actin network. The correlation time we observe in the curvature of actin-propelled bead paths may be a limiting time for the change in direction of a growing actin network. Biochemical events that attempt to change the direction of a growing actin network, such as chemotaxis or phagocytosis, must do this in the presence of an intrinsic curvature and temporal correlation like that described here. Future experiments on the spatial and temporal details of fluctuations and remodeling of actin network architectures will be needed to uncover the origin of these slow time scales.
Glossary
- Dendritic actin network
A network of biological polymers characterized by polarized Y-branches, where the pointed end of each filament is attached to the side of another filament and the barbed end grows forward
- Optical-force microscopy
A technique that uses the interaction of a focused laser beam with a micron-sized object to apply forces and monitor movements in 3D
- Frenet–Serret frame
The set of three orthogonal vectors, the tangent, normal and binormal that describes the local orientation of a point along a curve in 3D space
- Network debranching
The process by which a branched filament detaches from the side of another filament due to the hydrolysis of ATP in the Arp2 branching protein
Contributor Information
Joshua W Shaevitz, Email: shaevitz@princeton.edu.
Daniel A Fletcher, Email: fletch@berkeley.edu.
References
- 1.Welch MD, Mullins RD. Cellular control of actin nucleation. Annu Rev Cell Dev Biol. 2002;18:247–88. doi: 10.1146/annurev.cellbio.18.040202.112133. [DOI] [PubMed] [Google Scholar]
- 2.Pollard TD, Borisy G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–65. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 3.Fletcher DA, Theriot JA. An introduction to cell motility for the physical scientist. Phys Biol. 2004;1:T1–10. doi: 10.1088/1478-3967/1/1/T01. [DOI] [PubMed] [Google Scholar]
- 4.Gerbal F, Laurent V, Ott A, Carlier MF, Chaikin P, Prost J. Measurement of the elasticity of the actin tail of listeria monocytogenes. Eur Biophys J Biophys Lett. 2000;29:134–40. doi: 10.1007/s002490050258. [DOI] [PubMed] [Google Scholar]
- 5.Jeng RL, Goley ED, D’Alessio JA, Chaga OY, Svitkina TM, Borisy GG, Heinzen RA, Welch MD. A rickettsia wasp-like protein activates the arp2/3 complex and mediates actin-based motility. Cell Microbiol. 2004;6:761–9. doi: 10.1111/j.1462-5822.2004.00402.x. [DOI] [PubMed] [Google Scholar]
- 6.Cameron L, Svitkina T, Vignjevic D, Theriot JA, Borisy G. Dendritic organization of actin comet tails. Curr Biol. 2001;11:130–5. doi: 10.1016/s0960-9822(01)00022-7. [DOI] [PubMed] [Google Scholar]
- 7.Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–26. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gouin E, Egile C, Dehoux P, Villiers V, Adams J, Gertler F, Li R, Cossart P. The ricka protein of rickettsia conorii activates the arp2/3 complex. Nature. 2004;427:457–61. doi: 10.1038/nature02318. [DOI] [PubMed] [Google Scholar]
- 9.Mogilner A, Oster G. Force generation by actin polymerization: II. The elastic ratchet and tethered filaments. Biophys J. 2003;84:1591–605. doi: 10.1016/S0006-3495(03)74969-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerbal F, Chaikin P, Rabin Y, Prost J. An elastic analysis of listeria monocytogenes propulsion. Biophys J. 2000;79:2259–75. doi: 10.1016/S0006-3495(00)76473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlier MF, Wiesner S, Le Clainche C, Pantaloni D. Actin-based motility as a self-organized system: mechanism and reconstitution in vitro. C R Biol. 2003;326:161–70. doi: 10.1016/s1631-0691(03)00067-2. [DOI] [PubMed] [Google Scholar]
- 12.Soo FS, Theriot JA. Adhesion controls bacterial actin polymerization-based movement. Proc Natl Acad Sci. 2005;102:16233–8. doi: 10.1073/pnas.0507022102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcy Y, Prost J, Carlier MF, Sykes C. Forces generated during actin-based propulsion: a direct measurement by micromanipulation. Proc Natl Acad Sci. 2004;101:5992–7. doi: 10.1073/pnas.0307704101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parekh SH, Chaudhuri O, Theriot JA, Fletcher DA. Loading history determines the velocity of actin-network growth. Natl Cell Biol. 2005;7:1119–23. doi: 10.1038/ncb1336. [DOI] [PubMed] [Google Scholar]
- 15.Cameron L, Giardini P, Soo F, Theriot JA. Secrets of actin-based motility revealed by a bacterial pathogen. Natl Rev Mol Cell Biol. 2000;1:110–9. doi: 10.1038/35040061. [DOI] [PubMed] [Google Scholar]
- 16.Wiesner S, Helfer E, Didry D, Ducouret G, Lafuma F, Carlier M-F, Pantaloni D. A biomimetic motility assay provides insight into the mechanism of actin-based motility. J Cell Biol. 2003;160:387–98. doi: 10.1083/jcb.200207148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cameron LA, Robbins JR, Footer MJ, Theriot JA. Biophysical parameters influence actin-based movement, trajectory, and initiation in a cell-free system. Mol Biol Cell. 2004;15:2312–23. doi: 10.1091/mbc.E03-12-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacayo C, Theriot JA. Listeria monocytogenes actin-based motility varies depending on subcellular location: a kinematic probe for cytoarchitecture. Mol Biol Cell. 2004;15:2164–75. doi: 10.1091/mbc.E03-10-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auerbuch V, Loureiro J, Gertler F, Theriot JA, Portnoy D. Ena/vasp proteins contribute to listeria monocytogenes pathogenesis by controlling temporal and spatial persistence of bacterial actin-based motility. Mol Microbiol. 2003;49:1361–75. doi: 10.1046/j.1365-2958.2003.03639.x. [DOI] [PubMed] [Google Scholar]
- 20.Shenoy VB, Tambe DT, Prasad A, Theriot JA. A kinematic description of the trajectories of listeria monocytogenes propelled by actin comet tails. Proc Natl Acad Sci. 2007;104:8229–34. doi: 10.1073/pnas.0702454104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rutenberg AD, Grant M. Curved tails in polymerization-based bacterial motility. Phys Rev E. 2001;64:021904. doi: 10.1103/PhysRevE.64.021904. [DOI] [PubMed] [Google Scholar]
- 22.Cameron L, Footer MJ, van Oudenaarden A, Theriot JA. Motility of acta protein-coated microspheres driven by actin polymerization. Proc Natl Acad Sci. 1999;96:4908–13. doi: 10.1073/pnas.96.9.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaevitz JW, Fletcher DA. Load fluctuations drive actin network growth. Proc Natl Acad Sci. 2007;104:15688–92. doi: 10.1073/pnas.0702601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pralle A, Prummer M, Florin E, Stelzer EH, Horber JK. Three-dimensional high-resolution particle tracking for optical tweezers by forward scattered light. Micro Res Tech. 1999;44:378–86. doi: 10.1002/(SICI)1097-0029(19990301)44:5<378::AID-JEMT10>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 25.Lang MJ, Asbury CL, Shaevitz JW, Block SM. An automated two-dimensional optical force clamp for single molecule studies. Biophys J. 2002;83:491–501. doi: 10.1016/S0006-3495(02)75185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters I, de Grooth B, Schins J, Figdor C, Greve J. Three dimensional single-particle tracking with nanometer resolution. Rev Sci Instrum. 1998;69:2762–6. [Google Scholar]
- 27.Pralle A, Prummer M, Florin E-L, Stelzer EHK, Horber J. Three-dimensional high-resolution particle tracking for optical tweezers by forward scattered light. Microsc Res Tech. 1999;44:378–86. doi: 10.1002/(SICI)1097-0029(19990301)44:5<378::AID-JEMT10>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 28.Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of rna polymerase backtracking. Cell. 2003;115:437–47. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 29.Carlsson A. Growth velocities of branched actin networks. Biophys J. 2003;84:2907–18. doi: 10.1016/S0006-3495(03)70018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanchoin L, Pollard TD, Mullins RD. Interactions of adf/cofilin, arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr Biol. 2000;10:1273–82. doi: 10.1016/s0960-9822(00)00749-1. [DOI] [PubMed] [Google Scholar]
- 31.Zeile W, Zhang F, Dickinson R, Purich D. Listeria’s right-handed helical rocket-tail trajectories: mechanistic implications for force generation in actin-based motility. Cell Motil Cytoskeleton. 2005;60:121–8. doi: 10.1002/cm.20050. [DOI] [PubMed] [Google Scholar]
- 32.Robbins J, Theriot JA. Listeria monocytogenes rotates around its long axis during actin-based motility. Curr Biol. 2003;13:R754–6. doi: 10.1016/j.cub.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 33.Rafelski S, Theriot JA. Mechanism of polarization of listeria monocytogenes surface protein acta. Mol Microbiol. 2006;59:1262–79. doi: 10.1111/j.1365-2958.2006.05025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]