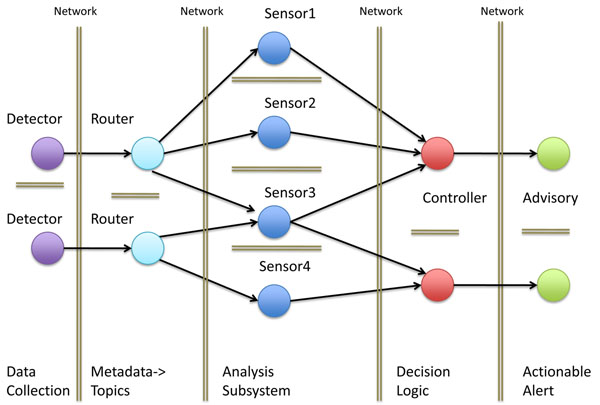
Figure 2.
A runtime view of the BioSITES system. BioSITES is achieved by connecting software components (the circles in the figure) from multiple institutions. These software components are connected via the message oriented middleware (MOM), ActiveMQ. MOM uses the publish-subscribe pattern to connect software components. Data is forwarded from one component to another when the second component subscribes to the first. BioSITES components include detectors (e.g. DNA sequencers and web crawlers), Routers (components that route data appropriately in the system based on the metadata), Sensors (analysis routines or workflows that identify and filter important signals in the data stream), Controllers (extensions of the Semantic Web that contain decision logic to process and integrate workflow outputs), and Advisories (components that contain reporting capabilities for displaying and relaying information). The double lines in the figure illustrate network boundaries. Data storage and access in the system is constrained by these network boundaries. Each component of the system is aware of all upstream components through messages that flow through the system.