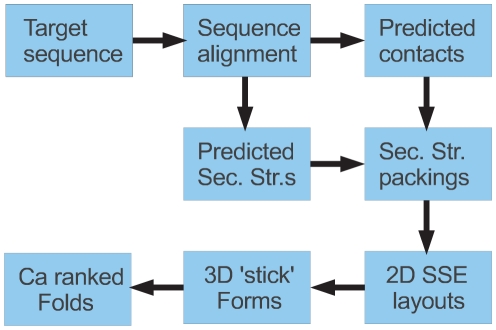
Figure 6. Overview of decoy model construction.
The Target sequence to be predicted is matched against the sequence database to generate a multiple Sequence alignment which is used both to predict secondary structure (Predicted Sec. Str.s) and residue contacts (Predicted contacts). These two derived data sets are combined to estimate pairwise packing interactions at the secondary structure element (SSE) level (Sec. Str. packings) which are used in the PLATO method firstly to select the structural class of the protein via 2D SSE layouts of the secondary structures. The corresponding stick models (3D ‘stick’ Forms) provide the framework over which different protein folds are combinatorially generated with pairings of secondary structures being evaluated by their predicted packing score. The ‘stick’ folds are then constructed at the residue (α-carbon) level giving the final set of Ca ranked Folds.