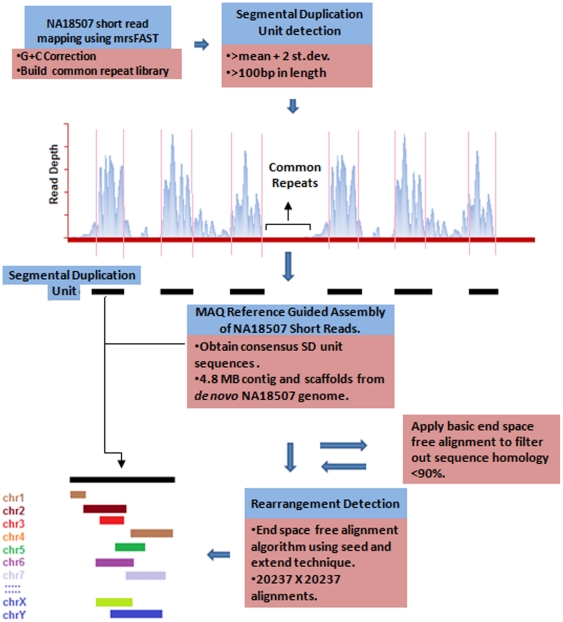
Figure 1. A schematic illustrating our hierarchical approach.
mrsFAST was used to obtain read depth distribution of the NA18507 human genome with maximum mismatch (n = 2) was allowed against the repeat masked reference human genome (build 36). A mean-based approach was utilized to computationally predict the boundaries of regions associated with excessive read depth. MAQ was used to obtain the consensus genome (mapping quality Q>30 and n = 2) from the NA18507 genome assembly. The consensus sequence for highly excessive read depth regions was obtained in order to apply a window-based alignment algorithm. The previously identified novel 4.8 Mb sequence from de novo assembly within this genome was also included in the rearrangement analysis.