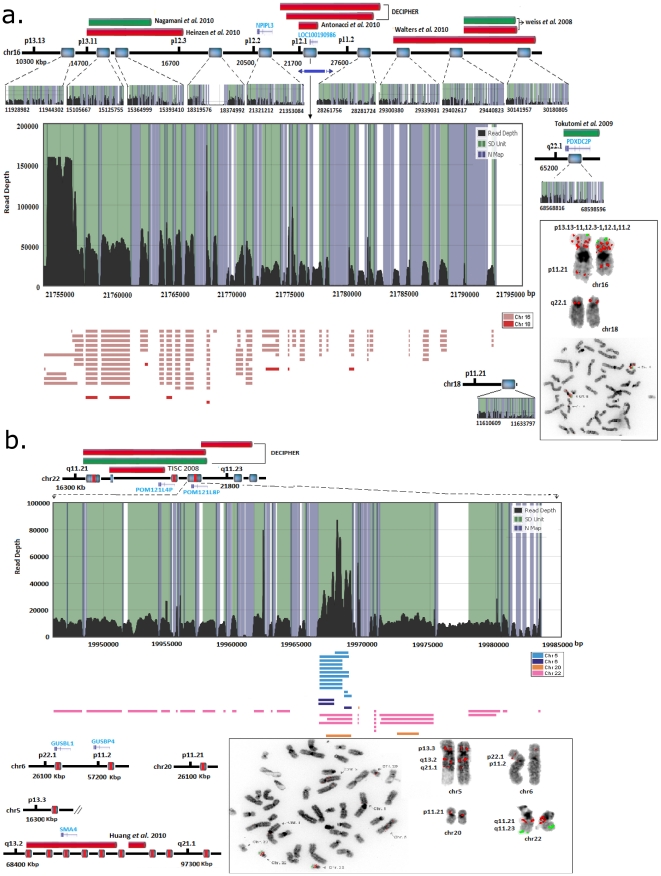
Figure 5. Signature of rearrangement hotspots located at a) 16p12.1 and b) 22q11.21.
A 40 kbp region within 16p12.1 is illustrated with its corresponding derivative copies which were localized by hierarchical analysis. This region consists of the NPIPL3 gene derivatives. The inter- and intra-chromosomal localization of the copies is approximated in the physical map within the chromosome contig (18p11.21). The alignments are color coded for chromosomes (i.e., color coded rectangles below the read depth plot) and FISH validation is illustrated for both inter- and intra-chromosomal localization. The pathogenic deletions and duplications located within these regions [27]–[32] are depicted in red and green bars, respectively The blue bars under the contig represent the approximated inversions previously reported by Antonacci, F. et al [26]. b) Analysis of a 37 kbp duplicated region within 22q11.21 revealed it is comprised of a core 2.7 kbp tandem duplicon copied from different chromosomes. Black lines represent the read depth (x-axis), green shade represent an SD unit, and blue bars represent the region with common repeat elements. The horizontal blocks (color coded according to chromosomes) are the rearrangement (intra/inter) fragments with >90% sequence similarity and >100 bp in length.