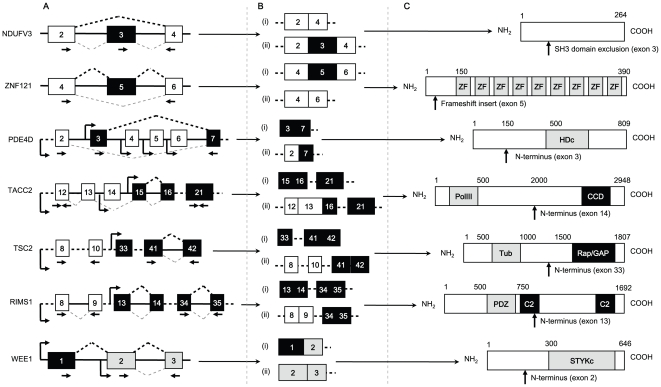
Figure 2. Overview of identified androgen-regulated mRNA isoforms.
(A) Cartoon of intron/exon structure of gene, showing the splicing pattern and the positions of the primers (shown as arrows) used for RT-PCR. Genes are annotated with the androgen responsive splicing/promoter selection event above the gene (exons joined by broken lines in black), and constitutive event below the gene (exons joined by broken lines in grey). The androgen-regulated exons are shown in black, and weakly androgen-dependent exons indicated in grey. Constitutive exons are indicated in white. The positions of the primers used for RT-PCR validation are shown below exons as arrows, and predicted position of alternative promoters are shown as angled arrow. (B) For each gene a pair of alternative mRNA transcript isoforms is shown. (C) The predicted full-length proteins made from each of the genes are indicated, along with the position of conserved domains. These are abbreviated: ZF = zinc finger motif; HDc = metal dependent phosphohydroxylate with conserved HD motif; PolIII = DNA polymerase subunits gamma and tau; CC = coiled coil domain; Tub = tuberin superfamily; Rap/GAP = RAP/GTPase Activating Protein superfamily; PDZ = protein interaction and binding domain; C2 = protein kinase C conserved domain 2 Ca2+ binding motif; STYKc = STYK protein kinase domain.