Cullin RING E3 ligases require covalent modification with Nedd8 for activity. Neddylation is reversed by the COP9 signalosome (CSN). We characterize the role of CSN-dependent deneddylation in vivo and propose a model in which CSN binds to cullin ligases in their active conformation and functions to recruit important regulatory factors.
Abstract
Cullin RING ligases (CRLs) are the largest family of cellular E3 ubiquitin ligases and mediate polyubiquitination of a number of cellular substrates. CRLs are activated via the covalent modification of the cullin protein with the ubiquitin-like protein Nedd8. This results in a conformational change in the cullin carboxy terminus that facilitates the ubiquitin transfer onto the substrate. COP9 signalosome (CSN)-mediated cullin deneddylation is essential for CRL activity in vivo. However, the mechanism through which CSN promotes CRL activity in vivo is currently unclear. In this paper, we provide evidence that cullin deneddylation is not intrinsically coupled to substrate polyubiquitination as part of the CRL activation cycle. Furthermore, inhibiting substrate-receptor autoubiquitination is unlikely to account for the major mechanism through which CSN regulates CRL activity. CSN also did not affect recruitment of the substrate-receptor SPOP to Cul3, suggesting it may not function to facilitate the exchange of Cul3 substrate receptors. Our results indicate that CSN binds preferentially to CRLs in the neddylation-induced, active conformation. Binding of the CSN complex to active CRLs may recruit CSN-associated proteins important for CRL regulation. The deneddylating activity of CSN would subsequently promote its own dissociation to allow progression through the CRL activation cycle.
INTRODUCTION
The COP9 signalosome (CSN) is an evolutionarily conserved complex consisting of eight subunits with similarity to the lid of the 26S proteasome regulatory particle (Cope and Deshaies, 2003; Wolf et al., 2003; Schwechheimer, 2004; Bosu and Kipreos, 2008; Wei et al., 2008). Loss of CSN activity as a result of deletion of different CSN subunits causes a constitutive photomorphogenic phenotype in plants and sterility in Caenorhabditis elegans and is lethal in Drosphila and mice (Chamovitz et al., 1996; Freilich et al., 1999; Yan et al., 2003; Orsborn et al., 2007; Dohmann et al., 2008). Lethality of CSN inactivation in mice is due to cell cycle defects. It has recently been shown that Cre recombinase–dependent knockout of CSN5 in mouse embryonic fibroblasts leads to multiple cell cycle defects and cell death (Yoshida et al., 2010), further emphasizing the essential role of CSN. Similarly, in Arabidopsis, CSN is essential for G2-phase progression and genomic stability (Dohmann et al., 2008).
The major function of CSN is the proteolytic cleavage of the isopeptide bond between the ubiquitin-like protein Nedd8 and cullin proteins (Lyapina et al., 2001; Cope et al., 2002; Schwechheimer, 2004). Cullins function as scaffold proteins for the assembly of cullin RING E3 ubiquitin ligases (CRLs) and constitute the largest family of cellular E3 ubiquitin ligases. All of the six well-characterized cullin proteins in mammalian cells (Cul1, Cul2, Cul3, Cul4a, Cul4b, and Cul5) bind via their C-termini to the small RING domain protein Rbx1 or Rbx2, which functions to recruit the ubiquitin-charged E2-conjugating enzyme. The cullin N-terminus interacts with cullin-specific substrate-receptor subunits, usually via adaptor proteins that mediate the interaction between cullin and substrate-receptor subunits (Petroski and Deshaies, 2005; Bosu and Kipreos, 2008).
CRLs require the covalent modification of a conserved C-terminal lysine residue with Nedd8. Neddylation activates CRLs by inducing a conformational change in the cullin C-terminus that results in increased flexibility of the Rbx1 RING domain, thus imparting multiple catalytic geometries to the E2-conjugating enzyme and promoting ubiquitin transfer onto the substrate (Duda et al., 2008; Saha and Deshaies, 2008; Yamoah et al., 2008). In addition, it has also been shown that cullin neddylation promotes the recruitment of ubiquitin-charged E2 enzyme (Kawakami et al., 2001; Sakata et al., 2007). Consequently, CSN-mediated cullin deneddylation leads to inhibition of CRL activity in vitro. In contrast to the negative regulation of CRL activity in vitro, there is clear evidence that CSN functions as a positive regulator of CRLs in vivo (Bosu and Kipreos, 2008; Wei et al., 2008). Thus loss of function of CSN leads to accumulation of CRL substrates and inhibition CRLs in vivo. A number of mechanisms for the positive role of CSN in regulating CRL activity have been proposed. It has been shown that CSN prevents the autoubiquitination of a number of cullin substrate-receptor subunits (Bosu and Kipreos, 2008; Schmidt et al., 2009; and references therein). This may be a consequence of cullin deneddylation and subsequent inactivation of CRLs in the absence of a bound substrate or of recruiting the CSN-associated deubiquitinating enzyme Ubp12/Usp15 to the CRL complex. Ubp12/Usp15 has been shown to function to reverse substrate-receptor autoubiquitination (Zhou et al., 2003; Hetfeld et al., 2005). Because both Nedd8 conjugation and deconjugation are required for CRL function, it has also been suggested that in vivo CRLs undergo rapid neddylation and deneddylation cycles (Cope and Deshaies, 2003). Based on this, a CRL activation cycle has been proposed in which substrate binding to the CRL complex induces cullin neddylation, thus leading to CRL activation. Upon substrate ubiquitination and dissociation, CSN-mediated deneddylation completes the activation cycle, which resumes when a new substrate binds.
In addition, CSN may function to regulate the binding of CAND1, a protein that interacts with all cullin homologues (Cope and Deshaies, 2003; Bosu and Kipreos, 2008). CAND1 interacts exclusively with unneddylated cullin proteins. Binding of CAND1 to cullins is mutually exclusive with the binding of substrate-receptor subunits. Thus, by deneddylating cullin proteins, CSN may function to promote CAND1 binding and consequently facilitate the exchange of substrate-receptor subunits (Lo and Hannink, 2006; Schmidt et al., 2009). Alternatively, CSN may compete with CAND1 for binding to cullin and thus prevent CAND1-mediated CRL disassembly. Finally, CSN has also been reported to play a regulatory role by promoting the dissociation of polyubiquitinated proteins from the CRL complex (Miyauchi et al., 2008).
In this study, we used a mammalian cellular system to investigate the importance of the various mechanisms of CSN-dependent CRL regulation. We also utilized MLN4924, a mechanism-based inhibitor of the Nedd8 E1-activating enzyme (Soucy et al., 2008; Brownell et al., 2010). On the basis of our findings, we propose a new model, in which CSN binding to CRLs and deneddylation are part of the neddylation cycle and regulate CRL activity.
RESULTS AND DISCUSSION
Determinants of substrate-receptor autoubiquitination
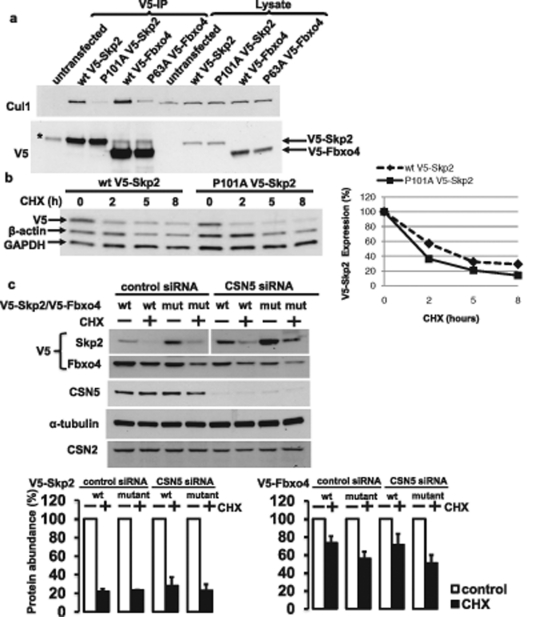
It has been suggested that CSN-mediated deneddylation is required to prevent the autoubiquitination of substrate receptors in the absence of a bound substrate. However, not all substrate receptors are subject to autoubiquitination. For the F-box protein family of Cul1 substrate receptors in fission yeast, it has recently been suggested that the presence of a proline residue in the N-terminal part of the F-box domain determines the affinity of the substrate receptor for Skp1 and Cul1 and the likelihood for CRL-mediated autoubiquitination (Schmidt et al., 2009). To determine the importance of the conserved proline residue in a mammalian system, we substituted the respective proline with alanine residues in a number of F-box proteins. As shown in Figure 1A, P101A Skp2 and P63A Fbxo4 showed markedly reduced Cul1 binding compared with the wild-type proteins. However, for Skp2, we observed no increase in the half-life of the P101A mutant compared with the wild-type protein under basal conditions (Figure 1B), suggesting that APCCdh1-mediated Skp2 ubiquitination but not autoubiquitination is normally the major mechanism through which Skp2 protein stability is regulated (Bashir et al., 2004; Wei et al., 2004). Even when CSN5 was knocked down, which is expected to increase autoubiquitination, the decrease in Skp2 protein expression after 8 h treatment with the protein synthesis inhibitor cycloheximide was similar for wild-type and P101A mutant Skp2 (Figure 1C). In contrast to Skp2, the proline mutant of Fbxo4 was degraded slightly faster under both basal and CSN5 knockdown conditions compared with the wild-type protein, which was relatively stable (Figure 1C). Indeed, when we treated cells with proteasome inhibitor MG-132, we noted the accumulation of polyubiquitinated mutant but not wild-type Fbxo4 (Supplemental Figure S1A). Given the markedly reduced binding of P63A Fbxo4 to Cul1, this ubiquitination is likely to be independent of the Cul1 CRL. Indeed, it was not significantly inhibited upon knockdown of Cul1 CRL adaptor protein Skp1, upon treatment with the Nedd8-activating enzyme (NAE) inhibitor MLN4924 (Soucy et al., 2008), and upon transfection of dominant-negative Cul1 (Figure S1, B and C).
FIGURE 1:
Role of the conserved proline residue in determining F-box protein binding to Cul1 and F-box protein stability. (A) HEK293T cells were transfected with the wild-type or proline mutant V5-Skp2 or V5-Fbxo4. Cell lysates were subjected to V5 immunoprecipitation and immunoprecipitates were analyzed by Western blotting with Cul1 and V5 antibodies. An asterisk (*) denotes a nonspecific band (heavy chain). (B) Cells were transfected with wild-type or P101A V5-Skp2 and subjected to chase analysis with 40 μM cycloheximide in which cells were lysed at the indicated time points after addition of the protein synthesis inhibitor. The relative amounts of wild-type and P101A V5-Skp2, determined by densitometry, are shown in the graph at the right. (C) Cells were transfected with negative control or CSN5 siRNA for 3 d and with wild-type or mutant V5-Skp2 or V5-Fbxo4 for the last 2 d. Cycloheximide (40 μM) was added where indicated 8 h before cell lysis and cells lysates were analyzed by Western blotting with the indicated antibodies. Bottom, quantification of the V5-Skp2 and V5-Fbxo4 abundance by densitometry. The results represent the average of two (V5-Skp2) or three (V5-Fbxo4) independent experiments.
We also determined the significance of the corresponding proline residue in the F-box protein β-TrCP. β-TrCP was subject to autoubiquitination under basal conditions based on an increase in protein expression upon MLN4924 treatment, while there was a smaller increase for Skp2 and no effect for Fbxo4 (Figure S2A). Alignment of the N-terminal part of the F-box domain for Skp2, Fbxw7α, Fbxo4, and β-TrCP (Figure S2B, according to Schmidt et al., 2009) shows the conserved proline residue in position 2 and indicates that, unlike other F-box proteins, β-TrCP contains four additional amino acids between the conserved LPx and EψxxxIxxxL sequences (where ψ corresponds to hydrophobic amino acids). The F-box–domain proline in position 2 in Skp2 (and presumably in other F-box proteins) is localized at the beginning of α-helix H1 in Skp2 and makes contact with Cul1 (Schulman et al., 2000; Zheng et al., 2002). In contrast, in the structure of the Skp1-β-TrCP complex, the proline residue is positioned in α-helix H0 and therefore presumably does not interact with Cul1 (Wu et al., 2002). Indeed, mutation of the proline to alanine (P185A β-TrCP) had no effect on the interaction of β-TrCP and Cul1 (Figure S2C). Based on the structure of the Skp1-β-TrCP complex, a likely more accurate alignment would place Leu189 and not Leu184 into the position of the conserved leucine residue in position 1 of the F-box domain (Figure S2C). To test for the importance of the two different leucine residues for binding to Cul1, we mutated both amino acids separately to lysine. However, both the L184K and L189K mutations in β-TrCP were without significant effect on the interaction between the two proteins (Figure S2C). In addition to the proline, the negatively charged residue in position 4 of the F-box is involved in the interaction with Cul1 (Zheng et al., 2002). However, this residue is absent in β-TrCP and is replaced by a histidine. We therefore tested whether the aspartate that precedes the histidine is involved in the binding of β-TrCP to Cul1. However, we observed that the D190A β-TrCP mutant also did not show reduced binding. Our results thus suggest that the N-terminal part of α-helix H1 in β-TrCP is not critically involved in the interaction of this F-box protein with Cul1 and that indirect contacts via Skp1 are likely sufficient to mediate Cul1 binding.
In conclusion, our results suggest that although the conserved F-box proline residue is important for binding of the F-box proteins Skp2 and Fbxo4 to Cul1, it is not the sole determinant of binding affinity and also does not appear to determine the rate of substrate-receptor autoubiquitination in mammalian cells. Based on our results and various other studies, it is evident that not all substrate receptors are subject to autoubiquitination. On the contrary, a number of substrate receptors, such as the Cul2 substrate receptor von Hippel-Lindau (VHL) protein and the Cul5 substrate receptor SOCS3, are stabilized upon incorporation into CRL complexes (Schoenfeld et al., 2000; Kamura et al., 2002; Haan et al., 2003), and this may also be true for Fbxo4 (see Figures 1C and S1). Interestingly, even though the VHL protein is not subject to Cul2 CRL-mediated autoubiquitination, knockdown of CSN2 or CSN5 in mammalian cells delays the CRL2VHL-dependent HIF-1α polyubiquitination and degradation (Miyauchi et al., 2008), suggesting that CSN is required for functions other than preventing autoubiquitination of substrate-receptor proteins. Two recent studies in Arabidopsis and Drosophila provide further support for this (Djagaeva and Doronkin, 2009; Spoel et al., 2009). In these studies, it was shown that the BTB (Broad-Complex, Tramtrack, and Bric a brac) proteins NPR1 and Kelch, respectively, bind to Cul3 and are subject to autoubiquitination. Importantly, lack of CSN activity in both Arabidopsis and Drosophila resulted in stabilization and elevation of the protein levels of the BTB substrate receptors (Djagaeva and Doronkin, 2009; Spoel et al., 2009), suggesting CSN does not inhibit autoubiquitination, but functions to promote CRL activity. In our further work, we therefore tested a number of hypotheses for a role of CSN in promoting CRL activity that is independent of substrate-receptor autoubiquitination prevention.
Hypothesis 1: CSN promotes CRL activity by mediating cycles of neddylation and deneddylation
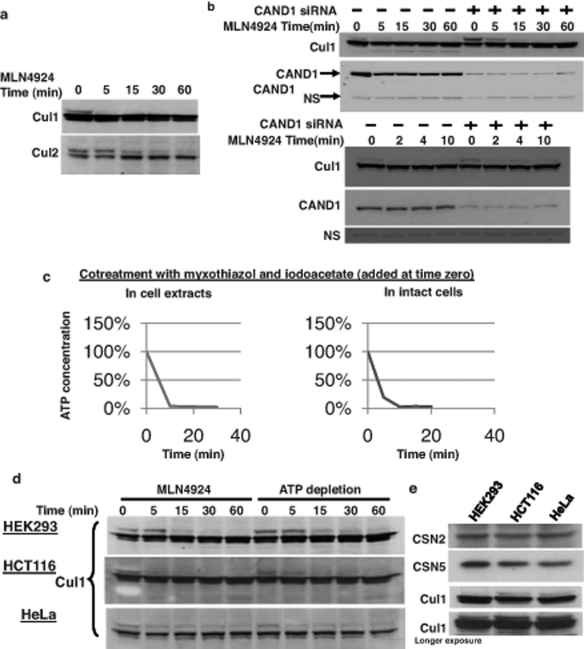
In vivo, both neddylation and deneddylation are required for efficient substrate ubiquitination. Based on this, it has been suggested that in vivo CRLs undergo rapid cycles of neddylation and deneddylation (Cope and Deshaies, 2003; Bosu and Kipreos, 2008; Wei et al., 2008). With the development of the specific inhibitor of cullin neddylation MLN4924, it has become possible to determine the in vivo deneddylation rates. As shown in Figure 2A, the deneddylation rates of endogenous Cul1 and Cul2 in HCT116 cells were relatively fast, demonstrating a marked decrease at 5 min after drug addition for Cul1 and at 15 min for Cul2, thus confirming results in the study by Soucy et al. (2009). The deneddylation rate of Cul1 in other cell lines was slightly slower (see Figure 2D), which is likely unrelated to the CSN expression level (Figure 2E). Given that it has been suggested that the CAND1 protein promotes cullin deneddylation (Min et al., 2005; Chew et al., 2007), we also determined the effect of small interfering RNA (siRNA)-mediated silencing of CAND1 on the deneddylation rate in HEK293 cells. As shown in Figure 2B, CAND1 silencing resulted in an increased basal Cul1 neddylation level, indicating that the knockdown of the CAND1 protein was functional. However, silencing of CAND1 was without effect on the rate of Cul1 deneddylation, even when the initial deneddylation rate was monitored during the first minutes after addition of MLN4924 (see Figure 2B, bottom). This suggests that CAND1 may not function to promote cullin deneddylation in vivo, possibly due to limiting expression levels relative to Cul1 (Chua et al., 2011).
FIGURE 2:
In vivo cullin deneddylation rates in the presence and absence of ongoing substrate ubiquitination. (A) HEK293 cells were grown in 12-well plates and 3 μM MLN4924 was added at time zero. Cells were lysed at the indicated time points and cell lysates were analyzed by Western blotting with Cul1 and Cul2 antibodies. (B) Cells were transfected with negative control or CAND1 siRNA for 3 d. The Cul1 deneddylation rate was determined as in (A). NS, nonspecific band that served as a loading control. (C) Cells were cotreated with 1 μM myxothiazol and 2.5 mM iodoacetate to rapidly deplete cellular ATP concentrations. ATP concentrations were measured as described in Materials and Methods. (D) To measure the Cul1 deneddylation rate in the presence and absence of ongoing substrate ubiquitination, HEK293, HCT116, and HeLa cells were treated with either MLN-4924 (3 μM) or myxothiazol (1 μM) and iodoacetate (2.5 mM) at time zero. Cells were lysed at the indicated time points and cell lysates were analyzed by Western blotting with Cul1 antibody. The Western blots shown are representative of at least three independent experiments in each cell line and did not show any consistent difference in the rate at which neddylation occurs. (E) The relative expression levels of CSN and Cul1 in HEK293, HCT116, and HeLa cells was compared by Western blotting of equal protein amounts of cell lysate with the indicated antibodies.
In its simplest form, the CRL activation cycle proposes that cullins are neddylated in the presence of a bound substrate and deneddylated after polyubiquitination and dissociation of the substrate (Cope and Deshaies, 2003; Bosu and Kipreos, 2008). Thus, if such CRL activation cycles operate in vivo, then it can be predicted that cullin deneddylation only occurs after the substrate has been polyubiquitinated. Hence, if substrate ubiquitination is inhibited, the rate of cullin deneddylation should be delayed. To test the hypothesis that cullin deneddylation is coupled to substrate ubiquitination, it was necessary to acutely inhibit ubiquitination. The only commercially available ubiquitination inhibitor, PYR-41, proved to be inefficient (unpublished data). As an alternative approach, we cotreated cells with the glycolysis inhibitor iodoacetate and the mitochondrial electron transport chain inhibitor myxothiazol to rapidly deplete the cellular ATP required for ubiquitination at the step of ubiquitin activation by the E1 enzyme. As shown in Figure 2B, combined treatment with iodoacetate and myxothiazol caused a rapid decline in the cellular free-ATP concentration, reaching virtually zero after 10 min. As expected, the treatment severely compromised cell viability and cells started to detach after about 1 h of treatment. However, since deneddylation occurs over a much shorter time course, we utilized this approach to determine whether substrate ubiquitination is a requirement for CSN-mediated cullin deneddylation. Thus we carried out experiments in which we compared the rate of Cul1 deneddylation in the presence of the specific neddylation inhibitor MLN4924 with that upon rapid ATP depletion, which leads to the inhibition of both neddylation and ubiquitination (which are both ATP dependent). We did not observe a significant difference in the deneddylation rate between the two different conditions in all tested cell lines (Figure 2D). Even when we preincubated the cells with iodoacetate and myxothiazol for 5 min prior to the addition of MLN4924 to ensure more ATP depletion during the deneddylation chase period, there was no decrease in the deneddylation rate in ATP-depleted cells (Figure S3). To further assess any role of substrate ubiquitination in regulating Cul1 deneddylation, we measured the deneddylation rate upon addition of MLN4924 in the presence of MG-132 to inhibit the degradation of polyubiquitinated proteins and under conditions of overexpression of the CRL E2-conjugating enzyme cdc34. However, none of these manipulations caused a change in the Cul1 deneddylation rate (Figure S4, A and B).
Taken together, our results suggest that cullin deneddylation is constitutive and not dependent on and coupled to substrate ubiquitination. While various reports have provided evidence that substrate binding induces cullin neddylation (Read et al., 2000; Bornstein et al., 2006; Sufan and Ohh, 2006; Chew and Hagen, 2007), our experiments suggest that cullin deneddylation is not linked to substrate ubiquitination in the CRL activation cycle. It is possible that an activation cycle involving only a smaller subpopulation of active CRL complexes exists in vivo. However, even when measuring the rate of deneddylation of Skp2-bound Cul1, which is presumably part of an active E3 ligase complex, no difference in the deneddylation rates when comparing ATP depletion and specific Nedd8 E1 inhibition was observed (unpublished data). We conclude that cullin neddylation is highly dynamic in vivo. Furthermore, deneddylation appears to be constitutive and independent of whether substrates are being polyubiquitinated. This suggests that there may be no direct link between substrate polyubiquitination and CRL activation cycles.
Hypothesis 2: CSN-mediated cullin deneddylation facilitates substrate-receptor exchange
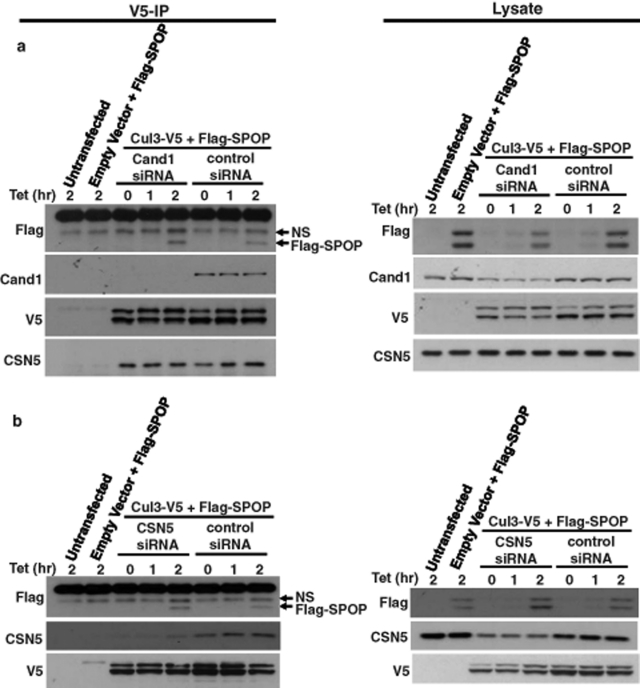
It has been proposed that CSN-mediated cullin deneddylation is necessary for efficient exchange of substrate-receptor modules (Lo and Hannink, 2006; Schmidt et al., 2009). According to this model, deneddylation promotes the binding of CAND1 to cullin proteins. CAND1 is known to interact only with unneddylated cullins. Furthermore, binding of substrate receptors and CAND1 to cullins is mutually exclusive. Thus binding of CAND1 would lead to the release of the substrate receptor. Subsequently, upon CAND1 dissociation, a new substrate-receptor module can be recruited to the cullin protein. This hypothesis has thus far been challenging to test due to the difficulty in measuring dynamic rates of substrate-receptor binding to and dissociation from cullins. We therefore planned to use a strategy based on rapidly introducing new substrate-receptor proteins into cells and measuring their rate of association with cullin proteins. However, various approaches, including transfection of recombinant substrate-receptor proteins and transduction using protein transduction domains, did not result in significant binding of the substrate-receptor proteins to cullins in cells. We therefore used a strategy based on induction of a substrate-receptor protein using a tetracycline-inducible expression system. Thus a plasmid encoding for the Cul3 substrate-receptor SPOP under control of a tetracycline-inducible promoter was transfected into HEK293 cells. SPOP expression was induced by addition of tetracycline and binding of the substrate receptors to Cul3 was determined by coimmunoprecipitation. As shown in Figure 3, the SPOP protein was induced upon tetracycline addition in a time-dependent manner and an obvious association between SPOP and Cul3 was observed after 2 h, as detected by Cul3-V5 immunoprecipitation. However, knockdown of CAND1 (Figure 3A) or CSN5 (Figure 3B) had no effect on the apparent rate with which SPOP bound to Cul3. This suggests that CAND1 binding to Cul3 upon CSN-mediated deneddylation is not required for the exchange of Cul3-bound substrate receptors with SPOP.
FIGURE 3:
Role of CSN-mediated cullin deneddylation in facilitating substrate-receptor exchange. Cells grown in 60-mm dishes were transfected with negative control or CAND1 siRNA, which was followed after 1 d by transfection of Cul3-V5 and FLAG-SPOP (in a tetracycline-inducible plasmid). Three days after siRNA transfection, 1 μg/ml tetracycline was added at time zero and cells were lysed at the indicated times; this was followed by V5 immunoprecipitation and Western blotting of immunoprecipitates and cell lysates with the indicated antibodies. (B) The experiment was performed as in (A), except that cells were transfected with negative control or CSN5 siRNA. NS, nonspecific band.
Hypothesis 3: CSN prevents CAND1-mediated CRL disassembly
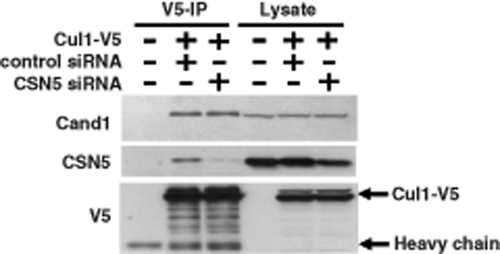
In contrast to the mechanism in hypothesis 2, in which CSN-mediated deneddylation promotes the binding of CAND1, it is also possible that the interaction of CSN with cullins prevents CAND1 binding and, subsequently, CAND1-mediated disassembly of the CRL complex. To test this hypothesis, we used siRNA to knock down the expression of CSN5 and measured the effect on the interaction between Cul1 and CAND1. As shown in Figure 4, silencing of CSN5 had no effect on the amount of CAND1 bound to Cul1. This suggests that CSN does not function to prevent binding of CAND1 to cullin proteins.
FIGURE 4:
Role of CSN in preventing CAND1-mediated CRL disassembly. Cells were transfected with negative control or CSN5 siRNA, which was followed after 1 d by transfection of Cul1-V5. Cells were lysed 3 d after siRNA transfection, and cell lysates were subjected to V5 immunoprecipitation and Western blotting with the indicated antibodies.
CSN binds preferentially to active CRLs
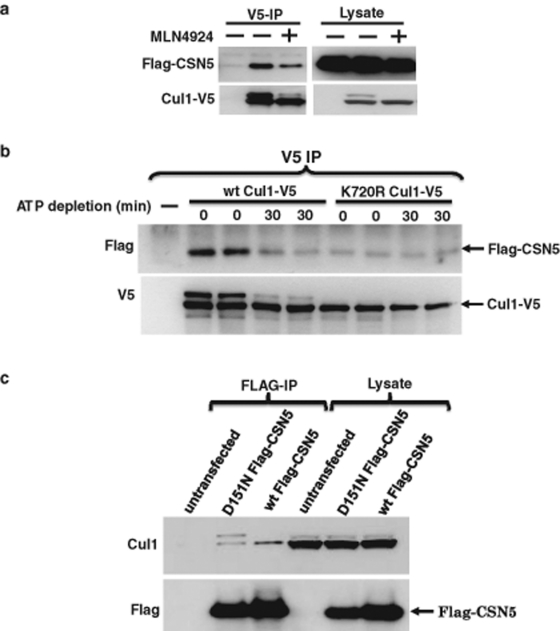
We finally considered the possibility that CSN binds to CRLs upon their activation by Nedd8. This may be a mechanism to recruit CSN-associated proteins that are important for CRL function. CSN-dependent cullin deneddylation may then terminate this process by causing the dissociation of the CSN complex from the CRL. To test this hypothesis, we determined whether cullin neddylation regulates CSN binding to cullin proteins in vivo. As shown in Figure 5A, inhibiting CRL neddylation with MLN4924 resulted in reduced CSN5 binding. Similarly, rapid depletion of cellular ATP (by cotreating cells with iodoacetate and myxothiazol, as described under Hypothesis 1) resulted in markedly reduced CSN5 binding to Cul1 (Figure 5B), consistent with reduced affinity of CSN for unneddylated Cul1. Furthermore, when we used the neddylation site mutant of Cul1 (K720R), basal binding of CSN5 was reduced (Figure 5B). Importantly, when using the K720R mutant there was no further decrease in CSN5 binding upon ATP depletion, indicating that the decrease in CSN5 binding to wild-type Cul1 is not due to nonspecific effects of the ATP depletion. We have previously reported that preventing binding of substrate receptors and substrates to Cul3 by using the S53A/F54A/E55A Cul3 mutant (which is unable to bind to BTB substrate-receptor proteins) causes a marked reduction in the neddylation level (Chew and Hagen, 2007). Consistent with our other results, reduced CSN5 binding to the mutant Cul3 was observed, compared with wild-type Cul3 (see Figure 6B).
FIGURE 5:
Preferential binding of CSN5 to neddylated Cul1. (A) Cells with stable expression of FLAG-CSN5 were transfected with Cul1-V5 and treated with 3 μM MLN-4924 for 3 h, as indicated. Cell lysates were used for V5 immunoprecipitation, which was followed by Western blotting. (B) Cells with stable expression of FLAG-CSN5 were transfected with wild-type or K720R mutant Cul1-V5 and subjected to 30 min of myxothiazol (1 μM) and iodoacetate (2.5 mM) treatment, indicated as ATP depletion, prior to cell lysis. Each condition in this experiment was performed in duplicate to ensure consistency of the results. (C) Cells were transfected with wild-type or D151N mutant FLAG-CSN5, which was followed by FLAG immunoprecipitation and Western blotting of immunoprecipitates and cell lysates with Cul1 and FLAG antibodies.
FIGURE 6:
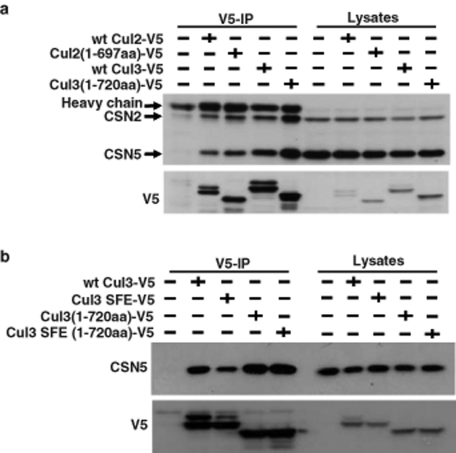
CSN5 binds preferentially to active CRLs. (A and B) HEK293T cells were transfected in 60-mm cell culture plates for 2 d with expression constructs for the full-length or extreme C-terminal deletion mutants of Cul2 and Cul3, as indicated at the top of each panel. The SFE mutant of Cul3 corresponds to the S53A/F54A/E55A mutant of Cul3, which is unable to bind to substrate-receptor subunits. The cells were lysed, and the lysates were subjected to V5 immunoprecipitation (IP), as described in Materials and Methods. Immunoprecipitates and aliquots of the cell lysates were analyzed by Western blotting with the indicated antibodies.
To confirm the preferential binding of CSN to neddylated cullins in our cellular system, FLAG-tagged CSN5 was immunoprecipitated from HEK293T cell lysates and binding of endogenous Cul1 was determined by Western blotting of immunoprecipitates (Figure 5C). Only unneddylated Cul1 was detected with wild-type CSN5. In contrast, a CSN mutant lacking deneddylation activity (D151N CSN5) coimmunoprecipitated approximately equal amounts of neddylated and unneddylated Cul1. Given that the neddylated form of Cul1 is far less abundant in the cell lysate, it can be concluded that CSN5 has a preference for binding to neddylated Cul1. The absence of neddylated Cul1 in wild-type CSN5 immunoprecipitates is likely due to deneddylating activity of CSN. These results are consistent with the hypothesis that cullin neddylation promotes binding of CSN.
We next wanted to determine whether the CSN binding is directly dependent on Nedd8 conjugation of cullins or whether it is an indirect consequence of the Nedd8-induced conformational change in the CRL C-terminus (Duda et al., 2008). To this end, we used extreme C-terminal deletion mutants of Cul2 and Cul3, which have previously been shown to not undergo neddylation and to harbor a constitutively active conformation able to support efficient substrate polyubiquitination, even in the absence of neddylation in vitro and in vivo (Duda et al., 2008; Yamoah et al., 2008; Boh et al., 2011). We observed that, compared with the full-length proteins, both C-terminally truncated Cul2 and Cul3 bound more CSN2 and CSN5, even though they lacked the modification with Nedd8 (Figure 6A). This suggests that it is not the conjugation with Nedd8 itself, but the Nedd8-induced conformational change in the CRL complex that may be important for CSN binding. To confirm the specificity of the increased binding of CSN2 and CSN5 to C-terminally truncated cullins, we mutated the conserved Lys441 and Arg442 residues in the four-helix bundle of Cul2 to prevent CSN binding to full-length cullin (Chua et al., 2011). Similarly, the K441E/R442E extreme C-terminal deletion mutant of Cul2 was unable to bind both CSN2 and CSN5 (Figure S5A) and lacked activity toward the Cul2 polyubiquitination target protein HIF-1α (Figure S5B).
It has been suggested that substrate polyubiquitination promotes CSN recruitment to the CRL complex (Miyauchi et al., 2008). Thus it is also possible that the observed preferential binding of the CSN complex to active CRLs is a consequence of increased amounts of bound polyubiquitinated substrates. To rule out this possibility, we introduced the S53A/F54A/E55A mutations into the extreme C-terminal deletion mutant of Cul3. These mutations in the interaction face of Cul3 with BTB proteins have been shown to prevent binding of substrate receptors and substrates to Cul3 (Chew and Hagen, 2007). As shown in Figure 6B, the S53A/F54A/E55A mutations in full-length Cul3 caused reduced CSN5 binding, which, as mentioned above, is likely due to reduced neddylation levels. In contrast, the Cul3 extreme C-terminal deletion mutant showed higher basal CSN5 binding that was not reduced upon introduction of the S53A/F54A/E55A mutations. Thus CSN binding to Cul3 is not directly dependent on substrate recruitment and polyubiquitination. We therefore conclude that the CSN complex preferentially binds to active CRL complexes.
Conclusion: role of CSN and CSN-mediated cullin deneddylation in the regulation of CRL activity
The mechanism through which CSN functions as a positive regulator of CRL activity in vivo is currently unclear. In this study, we provide evidence that CSN-mediated cullin deneddylation is not intrinsically coupled to substrate polyubiquitination in the CRL activation cycle. It has been proposed that CSN promotes CRL function indirectly by inhibiting substrate-receptor autoubiquitination and facilitating CAND1-mediated exchange of substrate-receptor subunits. Our results suggest that these may not be the exclusive functions of CSN. As an alternative function, CSN has been reported to promote the dissociation of polyubiquitinated substrates from the CRL complex in a manner that does not require deneddylation activity (Miyauchi et al., 2008). However, it was recently demonstrated by Yoshida et al., (2010) that the essential role of CSN in promoting cell cycle progression and cell survival is critically dependent on its deneddylation activity. This suggests that the dissociation of polyubiquitinated substrates is also unlikely to be the major function of CSN.
Our results indicate that CSN associates preferentially with active CRL complexes. This preferred binding may be due to a conformational change induced by the Nedd8 modification of the cullin protein. The recruited CSN complex may then exert functions important for CRL regulation. For instance, CSN may play a role in recruiting critical binding proteins to the CRL complex. CSN has been reported to associate with different protein kinases (Uhle et al., 2003) that may mediate important functions in the CRL activation cycle, such as the exchange of substrate-receptor subunits. The deneddylating activity of CSN may be important for promoting its own dissociation to allow progression through the CRL activation cycle. Thus studies on the role of CSN-binding proteins may be important to understand the mechanisms underlying the CRL activation cycle.
MATERIALS AND METHODS
Plasmid constructs
All plasmids used were previously described (Chew et al., 2007; Chew and Hagen, 2007; Boh et al., 2011), with the exception of the mammalian expression plasmid FLAG-SPOP and the retroviral expression plasmid for FLAG-CSN5. Both SPOP and CSN5 were amplified from MGC I.M.A.G.E. clones and subcloned into modified pcDNA3.1 with N-terminal FLAG tag. FLAG-CSN5 was subsequently transferred into the puro-MaRX retroviral expression vector (a kind gift from David Beach) and used to generate a stable FLAG-CSN5–expressing HEK293T cell line, as previously described (Gan et al., 2009). For DNA transfections, subconfluent cells were transfected using Genejuice (Novagen, San Diego, CA) according to the manufacturer's instructions.
siRNA-mediated gene silencing
For siRNA transfections, RNAi Max Lipofectamine (Invitrogen, Carlsbad, CA) was used as the transfection agent according to the manufacturer's instructions with the following annealed Silencer predesigned siRNA duplexes (Ambion, Austin, TX) at a final concentration of 25 nM: CAND1: siRNA ID 140584; CSN5: siRNA ID 214069; negative controls: Silencer negative control siRNA #2 or Mdm2 siRNA 122297. Cells were lysed 3 d after siRNA transfections for Western blot analysis.
Immunoblotting
For immunoblotting, the cells were washed with ice-cold phosphate-buffered saline and then lysed in Triton X-100-containing lysis buffer, as previously described (Culbert et al., 2001). Lysates were precleared by centrifugation before use for Western blotting. Equal amounts of protein were loaded for Western blot analysis. The following antibodies were used: rabbit polyclonal anti-Cul2 (51–1800; Zymed Laboratories (10672; Santa Cruz Biotechnology, Santa Cruz, CA), monoclonal anti-α-tubulin (236–10501; Molecular Probes, Invitrogen), monoclonal anti-V5 (AbD Serotec, Kidlington, UK), and monoclonal anti-FLAG M2 (Sigma-Aldrich, St. Louis, MO). Western blots shown are representative of at least two independent experiments.
Immunoprecipitation
V5 antibody (2.5 μg), coupled to 20 μl of protein G-Sepharose (Amersham Biosciences, GE Healthcare, Waukesha, WI), or 20 μl of FLAG-agarose (Sigma) was used for immunoprecipitations, and 500 μl of precleared lysate from HEK293T cells transfected in 60-mm tissue culture plates was added. The samples were tumbled at 4°C for 2 h, and the beads were then washed four times in 1 ml of NP40 cold lysis buffer (containing 50 mM NaCl, 0.5% NP-40, 5% glycerol, 0.5 mM EDTA, 50 mM Tris, pH 7.5, 1 mM dithiothreitol) and once in buffer containing 50 mM Tris (pH 7.5). The immunoprecipitated proteins were then denatured in SDS sample buffer and subjected to SDS–PAGE and Western blotting. The immunoprecipitation experiments shown are representative of at least two independent experiments.
ATP measurements
ATP measurement in intact cells using transfected firefly luciferase.
Cells in 12-well plates were transfected for 24 h with 0.2 μg of a firefly luciferase-pcDNA3 plasmid (under control of the cytomegalovirus promoter). Cells were trypsinized and resuspended in Krebs buffer. Intracellular ATP concentrations were determined by measuring the luminescence of aliquots of cells treated with myxothiazol and iodoacetate for various periods of time. Twenty microliters of the cells were then mixed with 100 μl of luciferase substrate, d-luciferin (Sigma) and used for ATP measurement.
ATP measurement in cell extracts.
Cells in 12-well plates were treated with myxothiazol and iodoacetate for different periods of time. Perchloric acid (6%) was used to extract the ATP from cells and the extract was spun for 1 min. The supernatant was collected and neutralized with 1.6 M K2CO3 containing 0.43 M triethanolamine buffer. The supernatant was spun for 1 min and used for ATP measurement by using the ENLITEN ATP assay system (Promega, Madison, WI).
Supplementary Material
Acknowledgments
We thank Jong-Bok Yoon (Yonsei University, Korea) for providing CAND1 cDNA, David Beach (Institute of Cell and Molecular Science, London) for providing the retroviral expression vector Puro-MaRX, and all the members of our laboratory for helpful discussions. This study was supported by grant number 07/1/21/19/500 from the Singapore Biomedical Research Council.
Abbreviations used:
- CRL
Cullin RING ligase
- CSN
COP9 signalosome
- NAE
Nedd8-activating enzyme
- siRNA
small interfering RNA
- VHL
von Hippel-Lindau
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-03-0251) on October 19, 2011.
REFERENCES
- Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M. Control of the SCFSkp2–Cks1 ubiquitin ligase by the APC/CCdh1 ubiquitin ligase. Nature. 2004;428:190–193. doi: 10.1038/nature02330. [DOI] [PubMed] [Google Scholar]
- Boh BK, Smith PG, Hagen T. Neddylation-induced conformational control regulates Cullin RING ligase activity in vivo. J Mol Biol. 2011;409:136–145. doi: 10.1016/j.jmb.2011.03.023. [DOI] [PubMed] [Google Scholar]
- Bornstein G, Ganoth D, Hershko A. Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate. Proc Natl Acad Sci USA. 2006;103:11515–11520. doi: 10.1073/pnas.0603921103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub JM, Matsui M, Deng XW. The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell. 1996;86:115–121. doi: 10.1016/s0092-8674(00)80082-3. [DOI] [PubMed] [Google Scholar]
- Chew EH, Hagen T. Substrate-mediated regulation of cullin neddylation. J Biol Chem. 2007;282:17032–17040. doi: 10.1074/jbc.M701153200. [DOI] [PubMed] [Google Scholar]
- Chew EH, Poobalasingam T, Hawkey CJ, Hagen T. Characterization of cullin-based E3 ubiquitin ligases in intact mammalian cells—evidence for cullin dimerization. Cell Signal. 2007;19:1071–1080. doi: 10.1016/j.cellsig.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Chua YS, Boh BK, Ponyeam W, Hagen T. Regulation of cullin RING E3 ubiquitin ligases by CAND1 in vivo. PLoS One. 2011;6:e6071. doi: 10.1371/journal.pone.0016071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope GA, Deshaies RJ. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114:663–671. doi: 10.1016/s0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- Culbert AA, Brown MJ, Frame SM, Hagen T, Cross DAE, Bax B, Reith AD. GSK-3 inhibition by adenoviral FRAT1 overexpression is neuroprotective and induces Tau dephosphorylation and β-catenin stabilisation without elevation of glycogen synthase activity. FEBS Lett. 2001;507:288–294. doi: 10.1016/s0014-5793(01)02990-8. [DOI] [PubMed] [Google Scholar]
- Dohmann EM, Levesque MP, De Veylder L, Reichardt I, Jürgens G, Schmid M, Schwechheimer C. The Arabidopsis COP9 signalosome is essential for G2 phase progression and genomic stability. Development. 2008;135:2013–2022. doi: 10.1242/dev.020743. [DOI] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djagaeva I, Doronkin S. COP9 limits dendritic branching via Cullin3-dependent degradation of the actin-crosslinking BTB-domain protein Kelch. PLoS One. 2009;4:e7598. doi: 10.1371/journal.pone.0007598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freilich S, Oron E, Kapp Y, Nevo-Caspi Y, Orgad S, Segal D, Chamovitz DA. The COP9 signalosome is essential for development of Drosophila melanogaster. Curr Biol. 1999;9:1187–1190. doi: 10.1016/S0960-9822(00)80023-8. [DOI] [PubMed] [Google Scholar]
- Gan FF, Chua YS, Scarmagnani S, Palaniappan P, Franks M, Poobalasingam T, Bradshaw TD, Westwell AD, Hagen T. Structure-activity analysis of 2′-modified cinnamaldehyde analogues as potential anticancer agents. Biochem Biophys Res Commun. 2009;387:741–747. doi: 10.1016/j.bbrc.2009.07.104. [DOI] [PubMed] [Google Scholar]
- Haan S, Ferguson P, Sommer U, Hiremath M, McVicar DW, Heinrich PC, Johnston JA, Cacalano NA. Tyrosine phosphorylation disrupts elongin interaction and accelerates SOCS3 degradation. J Biol Chem. 2003;278:31972–31979. doi: 10.1074/jbc.M303170200. [DOI] [PubMed] [Google Scholar]
- Hetfeld BK, Helfrich A, Kapelari B, Scheel H, Hofmann K, Guterman A, Glickman M, Schade R, Kloetzel PM, Dubiel W. The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Curr Biol. 2005;15:1217–1221. doi: 10.1016/j.cub.2005.05.059. [DOI] [PubMed] [Google Scholar]
- Kamura T, Brower CS, Conaway RC, Conaway JW. A molecular basis for stabilization of the von Hippel-Lindau (VHL) tumor suppressor protein by components of the VHL ubiquitin ligase. J Biol Chem. 2002;277:30388–30393. doi: 10.1074/jbc.M203344200. [DOI] [PubMed] [Google Scholar]
- Kawakami T, et al. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 2001;20:4003–4012. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SC, Hannink M. CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Mol Cell Biol. 2006;26:1235–1244. doi: 10.1128/MCB.26.4.1235-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- Min KW, Kwon MJ, Park HS, Park Y, Yoon SK, Yoon JB. CAND1 enhances deneddylation of CUL1 by COP9 signalosome. Biochem Biophys Res Commun. 2005;334:867–874. doi: 10.1016/j.bbrc.2005.06.188. [DOI] [PubMed] [Google Scholar]
- Miyauchi Y, Kato M, Tokunaga F, Iwai K. The COP9/signalosome increases the efficiency of von Hippel-Lindau protein ubiquitin ligase-mediated hypoxia-inducible factor-α ubiquitination. J Biol Chem. 2008;283:16622–16631. doi: 10.1074/jbc.M710599200. [DOI] [PubMed] [Google Scholar]
- Orsborn AM, Li W, McEwen TJ, Mizuno T, Kuzmin E, Matsumoto K, Bennett KL. GLH-1, the C. elegans P granule protein, is controlled by the JNK KGB-1 and by the COP9 subunit CSN-5. Development. 2007;134:3383–3392. doi: 10.1242/dev.005181. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Read MA, et al. Nedd8 modification of Cul-1 activates SCFβTrCP-dependent ubiquitination of IĸBα. Mol Cell Biol. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata E, Yamaguchi Y, Miyauchi Y, Iwai K, Chiba T, Saeki Y, Matsuda N, Tanaka K, Kato K. Direct interactions between NEDD8 and ubiquitin E2 conjugating enzymes upregulate cullin-based E3 ligase activity. Nat Struct Mol Biol. 2007;14:167–168. doi: 10.1038/nsmb1191. [DOI] [PubMed] [Google Scholar]
- Schmidt MW, McQuary PR, Wee S, Hofmann K, Wolf DA. F-box-directed CRL complex assembly and regulation by the CSN and CAND1. Mol Cell. 2009;35:586–597. doi: 10.1016/j.molcel.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld AR, Davidowitz EJ, Burk RD. Elongin BC complex prevents degradation of von Hippel-Lindau tumor suppressor gene products. Proc Natl Acad Sci USA. 2000;97:8507–8512. doi: 10.1073/pnas.97.15.8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C. The COP9 signalosome (CSN): an evolutionary conserved proteolysis regulator in eukaryotic development. Biochim Biophys Acta. 2004;1695:45–54. doi: 10.1016/j.bbamcr.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–737. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell. 2009;137:860–872. doi: 10.1016/j.cell.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sufan RI, Ohh M. Role of the NEDD8 modification of Cul2 in the sequential activation of ECV complex. Neoplasia. 2006;8:956–963. doi: 10.1593/neo.06520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhle S, et al. Protein kinase CK2 and protein kinase D are associated with the COP9 signalosome. EMBO J. 2003;22:1302–1312. doi: 10.1093/emboj/cdg127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Serino G, Deng XW. The COP9 signalosome: more than a protease. Trends Biochem Sci. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG Jr. Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194–198. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- Wolf DA, Zhou C, Wee S. The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nat Cell Biol. 2003;5:1029–1033. doi: 10.1038/ncb1203-1029. [DOI] [PubMed] [Google Scholar]
- Wu K, Chen A, Tan P, Pan ZQ. The Nedd8-conjugated ROC1-CUL1 core ubiquitin ligase utilizes Nedd8 charged surface residues for efficient polyubiquitin chain assembly catalyzed by Cdc34. J Biol Chem. 2002;277:516–527. doi: 10.1074/jbc.M108008200. [DOI] [PubMed] [Google Scholar]
- Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, Pan ZQ. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin1's C-terminal tail. Proc Natl Acad Sci USA. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Walz K, Nakamura H, Carattini-Rivera S, Zhao Q, Vogel H, Wei N, Justice MJ, Bradley A, Lupski JR. COP9 signalosome subunit 3 is essential for maintenance of cell proliferation in the mouse embryonic epiblast. Mol Cell Biol. 2003;23:6798–6808. doi: 10.1128/MCB.23.19.6798-6808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Yoneda-Kato N, Panattoni M, Pardi R, Kato J. YCSN5/Jab1 controls multiple events in the mammalian cell cycle. FEBS Lett. 2010;584:4545–4552. doi: 10.1016/j.febslet.2010.10.039. [DOI] [PubMed] [Google Scholar]
- Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, Zhang H. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell. 2002;10:1519–1526. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- Zhou C, Wee S, Rhee E, Naumann M, Dubiel W, Wolf DA. Fission yeast COP9/signalosome suppresses cullin activity through recruitment of the deubiquitylating enzyme Ubp12p. Mol Cell. 2003;11:927–938. doi: 10.1016/s1097-2765(03)00136-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.