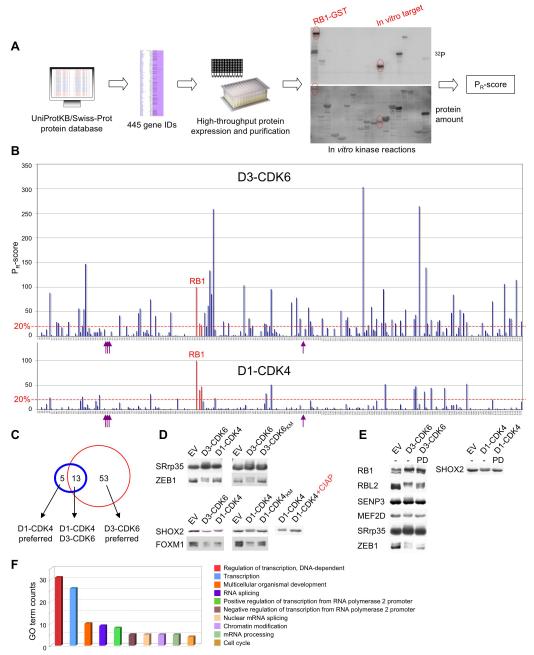
Figure 1. Proteome-Wide Identification of CDK4/6 Substrates.
(A) Schematic for the approach to substrate identification. All nuclear proteins with at least two potential CDK phosphorylation sites were selected from the protein database SWISS-PROT. Candidate proteins were expressed in E. coli, purified and tested for phosphorylation by recombinant cyclin D1-CDK4 and cyclin D3-CDK6. Reaction products were analyzed by SDS-PAGE (lower panel) and autoradiography (upper panel), and relative rates of phosphorylation (PR-scores) were calculated.
(B) Side-by-side comparison of phosphorylation of 285 enriched proteins by cyclin D3-CDK6 (upper panel) versus cyclin D1-CDK4 (lower panel). PR-scores are the rates of phosphorylation normalized to RB1, which was set to 100%. Proteins with PR-scores above 20% are referred to as in vitro substrates. Arrows indicate histone proteins; the three RB family proteins are highlighted in red.
(C) Overlap between cyclin D1-CDK4 (blue circle) and cyclin D3-CDK6 (red circle) in vitro substrates.
(D) In vitro substrates were co-expressed in HEK293 cells with either empty vector (EV) or wild-type cyclin-CDK complexes (left panel). Co-expression of wild type versus kinase-dead (KM) CDK versions is shown in the middle panel. Phosphorylated SHOX2 was treated with calf intestinal phosphatase (CIAP, right panel).
(E) Collapse in the mobility shift of seven CDK4/6 substrates upon short-term treatment (40 min) with the CDK4/6 specific inhibitor, PD0332991. GST-tagged substrates plus cyclin D-CDK complexes were expressed in HEK293 cells exposed to either 1 μM of the inhibitor (PD) or DMSO (−) 30 h post-transfection.
(F) ‘Biological process’ gene ontology (GO) terms were assigned to all 71 in vitro substrates. See also Figure S1 and Table S1.