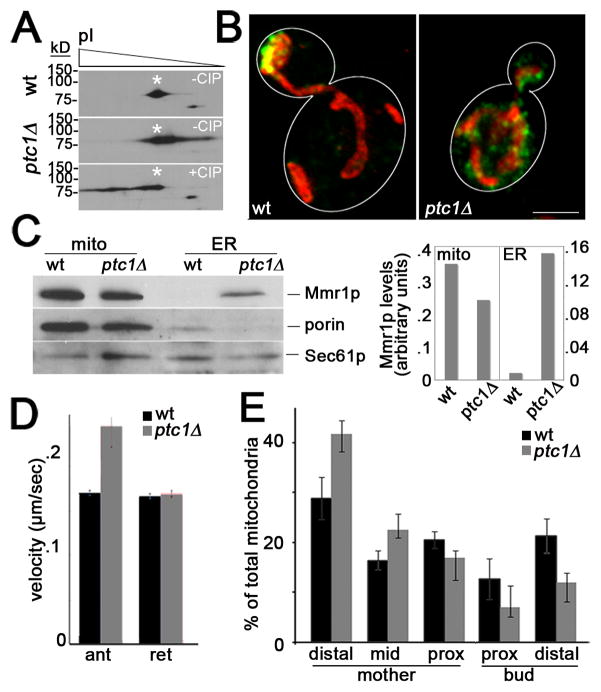
Fig. 4. Deletion of PTC1 results in mislocalization of Mmr1p, increased Mmr1p phosphorylation, increased velocity of anterograde mitochondrial movement and defects in anchorage of mitochondria in the bud tip.
(A) Mitochondria were isolated from wild-type cells (CZY057) or ptc1Δ cells (CZY106), each expressing myc-tagged Mmr1p, were analyzed by 2D gel electrophoresis. Deletion of PTC1 results in a shift of the isoelectric point of Mmr1p, consistent with increased phosphorylation of the protein. Treatment of mitochondrial extracts from ptc1Δ cells with calf alkaline phosphatase (CIP), as described in Fig. S4, dephosphorylates and alters the isoelectric point of Mmr1p. (B) Maximum projection of mitochondria (red) and Mmr1p (green) in a wild-type and a ptc1Δ cell. Cell outlines are shown in white. Bar, 1 μm. (C) Mitochondria and ER were isolated from wild-type (CZY057) and ptc1Δ (CZY106) cells expressing myc-tagged Mmr1p, as for Fig. 4. Left panel: The recovery of myc-tagged Mmr1p, mitochondrial (porin) and ER (Sec61p) marker proteins in fractions enriched in mitochondria (mito) and ER was determined as for Fig. 3C. Right panel: Quantitation of Mmr1p recovered in subcellular fractions. Values shown are arbitrary units normalized to recovery of a mitochondrial marker (porin) or an ER marker (Sec61p) for mitochondria and ER fractions, respectively. (D) ptc1Δ cells have increased anterograde mitochondrial velocity. The velocity of anterograde (ant) and retrograde (ret) mitochondrial movement in the mother cell and bud of wild-type (CZY001) and ptc1Δ (CZY089) cells was measured as for Fig. 2. Error bars are standard error of the mean. n >100 cells for each strain. * p<0.002. (E) Mitochondrial distribution in wild-type and ptc1Δ cells was determined as for Fig. 2.