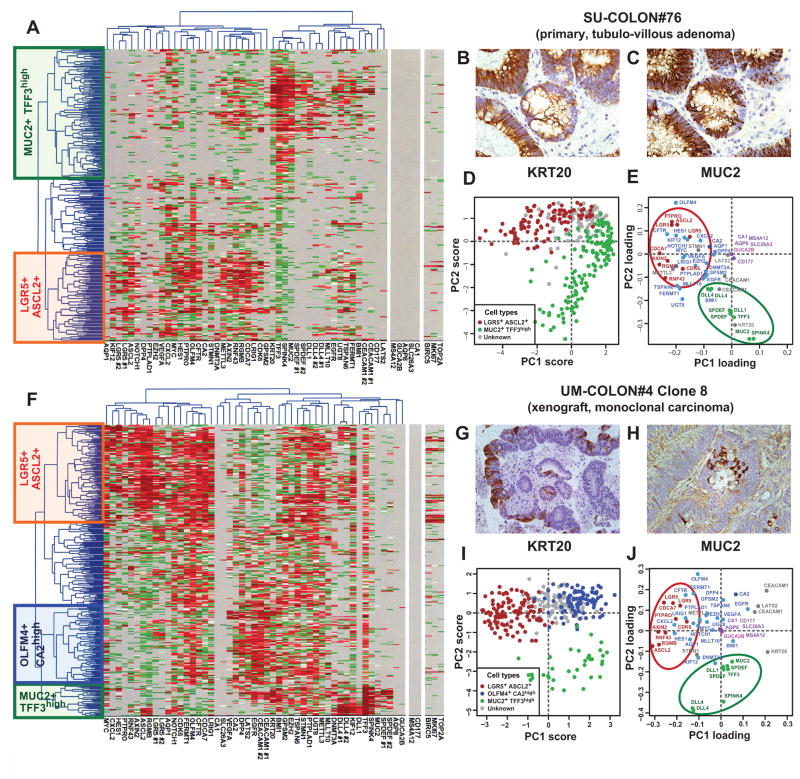
Figure 2. SINCE-PCR analysis of human colon tumor tissues.
Analysis by SINCE-PCR of the EpCAM+/CD44+ population from human colon tumor tissues was performed on a large primary benign adenoma (A, SU-COLON#76) and a monoclonal colon cancer xenograft obtained from injection of a single-cell (n = 1) in a NOD/SCID/IL2Rγ−/− mouse (F, UM-COLON#4 Clone 8). The analysis revealed the presence of multiple cell populations characterized by distinct gene signatures, closely mirroring lineages and differentiation stages observed in the EpCAM+/CD44+ population from the normal colon epithelium. Principal component analysis (PCA) of SINCE-PCR data confirmed hierarchical clustering results, visualizing distinct cell populations characterized by the coordinated expression of independent sets of genes that corresponded to those observed in normal tissues (D–E, SU-COLON#76; I–J, UM-COLON#4 Clone8). Most importantly, the analysis of the monoclonal tumor xenograft obtained from a single-cell indicated that these distinct cell populations, together with their distinctive gene-expression repertoires, did not arise as the result of the coexistence within the tumor tissue of independent genetic sub-clones, but as the result of multi-lineage differentiation processes during tumor growth. SINCE-PCR data were confirmed at the protein level by immunohistochemistry, testing for expression of KRT20 and MUC2 on corresponding tissue sections (B–C, SU-COLON#76; G–H, UM-COLON#4 Clone 8). Color coding of normalized Ct values in hierarchical clustering plots (A, F) and of gene-families in PC loading plots (E–J) are identical to Fig. 1.