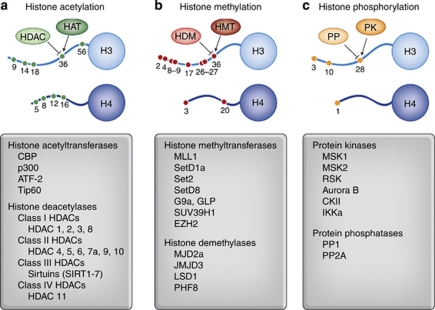
Figure 1.
Summary of well-understood histone modifications and histone-modifying enzymes. (a) Histone acetylation at numerous lysine residues on histone tails is catalyzed by histone acetyltransferases (HATs) and removed by histone deacetlyases (HDACs). Histone acetylation is generally a transcriptionally permissive mark. Different HAT and HDAC enzymes are listed below. Importantly, specific HDACs isoforms are differentially expressed across brain structures and appear to uniquely regulate different aspects of cognition. (b) Histone methylation at lysine and arginine residues on histone tails is catalyzed by histone methyltransferases (HMTs) and removed by histone demethylases (HDMs). Histone methylation at different amino acid residues has been linked to both transcriptional activation and transcriptional repression. Methylation can occur in mono-, di-, or even tri-methylated states. Many HDMs and HMTs are specific for modifications at individual amino acids on histone tails or even a specific number of methyl groups. (c) Histone phosphorylation at serine residues is catalyzed by protein kinases (PKs) such as mitogen- and stress-activated protein kinase 1 (MSK1), whereas phosphorylation marks are removed by protein phosphatases such as protein phosphatase 1 (PP1). Histone phosphorylation is generally linked to transcriptional activation.