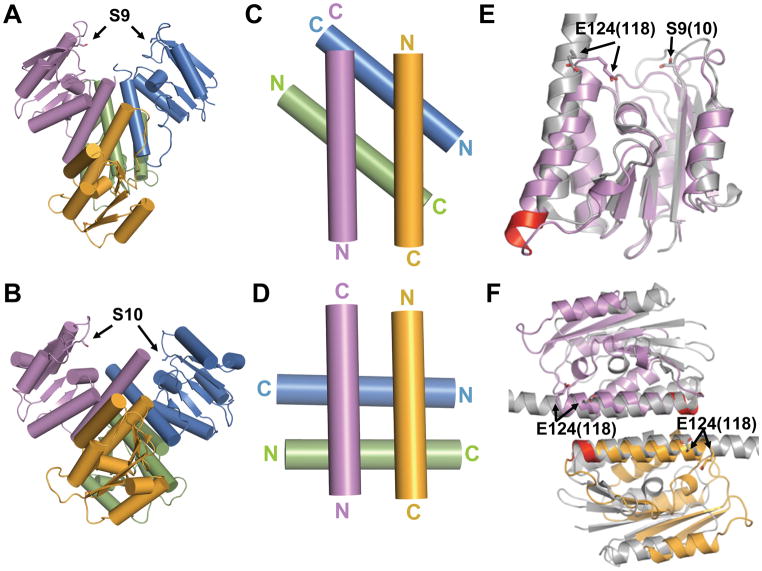
Figure 3. Comparison of the Sin and γδ activated tetramers.
A. Quaternary structure of Sin Q115R, shown as a cartoon with cylindrical helices.
B. Quaternary structure of γδ resolvase. The catalytic domain of a postcleavage structure of an activated mutant of γδ resolvase (1ZR4) is shown from the same vantage point as Sin in part A.
C. E helices of Sin. Only the E helices at the center of the activated tetramer are displayed. The two pairs cross at an angle of ~50°.
D. E helices of γδ resolvase. Only the E helices from the activated γδ resolvase catalytic domain structure (2GM5) are shown. In all the activated γδ resolvase structures, these cross at ~85°.
E. Comparison of activated Sin and γδ protomers. One protomer from each activated tetramer is superimposed, guided by the core region. Sin is in purple and γδ (1ZR4) gray. The active site serines and the conserved glutamate from helix E are shown as sticks (Sin E124 and γδ E118). Helix E of Sin has a 1-turn extension at its N-terminus that is highlighted in red. The angle between the axes of the two E helices is ~20°, and the difference in rotation about the helical axes is ~25 °.
F. Comparison of Sin and γδ rotating dimers. One rotating dimer from each activated tetramer was superimposed by aligning the approximate twofolds between the E-helices. Sin is colored as in part A, and γδ gray (1ZR4). (See also figure S2).