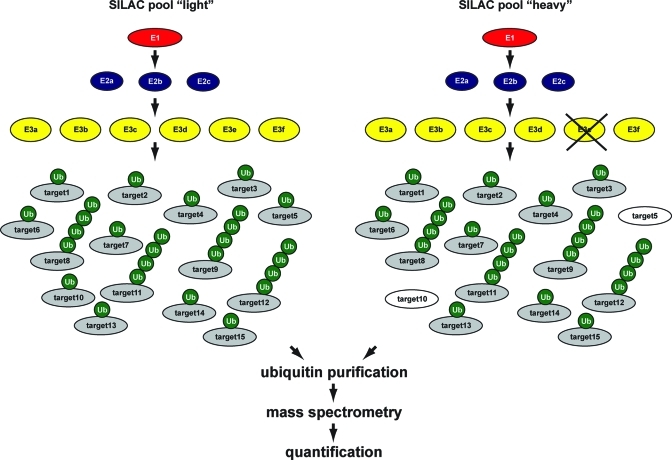
Figure 4.
Quantitative proteomics strategy to identify target proteins for ubiquitin E3 enzymes. SILAC technology is used for differential labeling of cells to generate two distinct pools. Using RNAi or specific inhibitors, E3 enzymes can be inactivated. Target proteins that are regulated by the indicated E3 enzyme can be identified by purifying ubiquitin from both pools of cells and analyzing the purified samples by mass spectrometry. Decreases in SILAC ratios for specific proteins upon E3 inactivation will reveal potential target proteins for this E3 enzyme. Similar strategies can be used to identify target proteins for ubiquitin proteases or ubiquitin proteome dynamics in response to specific stimuli. A third SILAC pool can be added as a negative control. Alternative quantitative proteomics methodologies are also available.