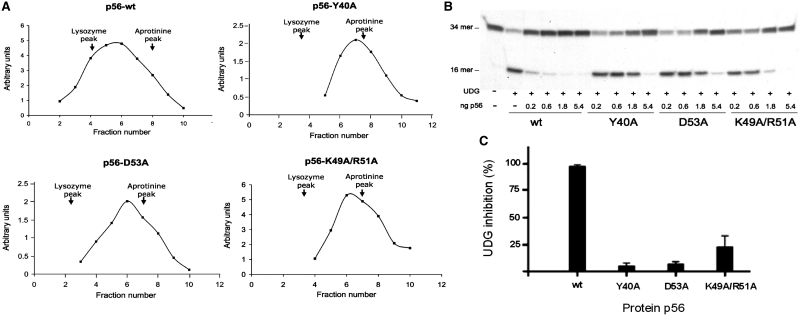
Figure 4.
Influence of selected residues on the stability of the p56 dimer and its inhibition activity. (A) The oligomerization state of p56 mutants was analysed by glycerol gradient sedimentation. Wild-type p56 and mutants, Y40A, D53A and K49A/R51A (12 µg of each), were loaded on a 15–30% glycerol gradient and subjected to centrifugation for 49 h at 59 000 rpm in a Beckman SW.65 rotor. As markers, 15 µg of lysozyme (14.7 kDa) and 15 µg of aprotinine (6.5 kDa) were loaded in the same gradient. Sedimentation was from right to left. After fractionation, aliquots from each fraction were analysed by SDS–PAGE. Densitometry scanning of the gels stained with Coomassie Blue was used to determine the amount of the different proteins in each fraction (arbitrary units). The arrows indicate fractions at which the maximal amount of each marker was detected. (B) Inhibition activity of p56 dimerization mutants. Increasing amounts of the p56 mutants (from 0.2 to 5.4 ng) were incubated with 5 pg of UDG. Then, the DNA substrate was added and incubated for 20 min at 37°C and the product was analysed in 8 M urea–20% polyacrylamide gels. (C) Bands corresponding to DNA substrate and DNA product were quantified by densitometry and the percentage of inhibition was calculated taking the wild-type p56 values as 100% inhibition. Data are depicted in a bar chart and average values of three independent experiments with standard deviations are represented.