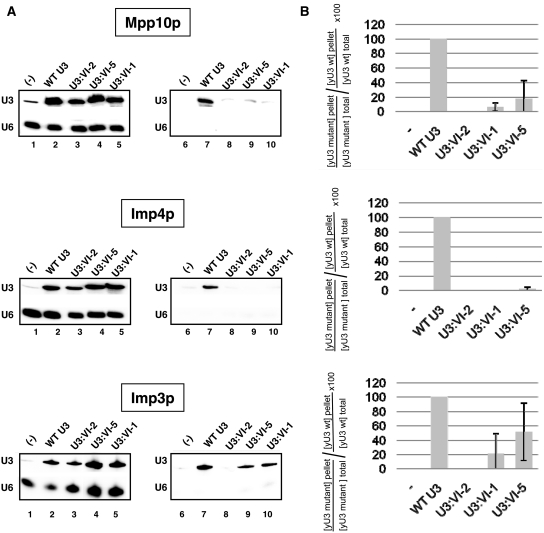
Figure 5.
Segment VI of U3 snoRNA is required for efficient association of proteins Mpp10, Imp4 and Imp3 in cellulo. (A) The JH84-SV10 cells (expressing the Flag-tagged Mpp10 protein and transformed with plasmid p413TEF::HA-Imp4p) and the JH84-SV3 cells (expressing the TAP-tagged Imp3 protein) were transformed with the WT or a variant pASZ11::U3A plasmid (U3:WT, U3:VI-1, U3:VI-2 and U3:VI-5). Cells were grown in YPG medium until stationary phase and then transferred in YPD medium for 24 h. They were washed with ice-cold water and lysed as previously described (20). A fraction of the extract (10%) was used to quantify the cellular amount of variant yU3A RNAs by northern blot analysis, using the same conditions as in Figure 2C (lanes 1–5). Another fraction of the extract (90%) was used for immunoselection assays, by incubation with beads coated either with an anti-Flag, an anti-HA antibody or with IgG as described in ‘Materials and Methods’ section. In both cases, the RNAs bound to the beads were extracted by proteinase K digestion followed by phenol extraction. The amount of the yU3A variant RNAs associated with the Flag-tagged Mpp10, HA-tagged Imp4 or TAP-tagged Imp3 protein was analysed by northern blot analysis as described above (Lanes 6–10). (B) The relative amounts of immunoselected variant yU3A RNAs, as compared to WT yU3A RNA were determined (IV/IWT%). As described in ‘Materials and Methods’ section, the binding capacity of the Flag-tagged Mpp10, HA-tagged Imp4 and TAP-tagged Imp3 proteins to the variant RNAs were expressed as a percentage of their binding capacity to the WT RNA (Flag-tagged Mpp10p, HA-tagged Imp4p and TAP-tagged Imp3p relative affinities indicated as 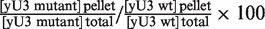 percentages). The values given are mean values of three independent experiments and their standard deviations are represented by bars.
percentages). The values given are mean values of three independent experiments and their standard deviations are represented by bars.