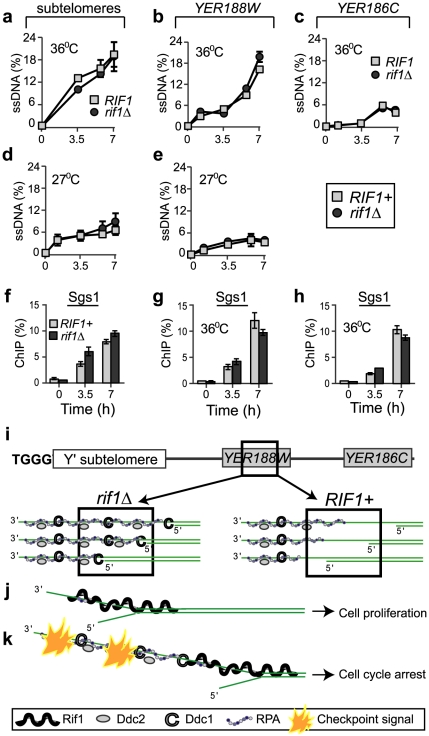
Figure 3. Rif1 does not inhibit resection of sub-telomeres and single gene loci.
(a–c) Quantification of ssDNA generated in cdc13-1 cells with a rif1Δ mutation (dark circles) or with wild-type RIF1 (light squares) at 36°C at: (a) 1 kb, in sub-telomeres, (b) 8 kb, in YER188W and (c) 15 kb, in YER186C. (d–e) As in a–b, except that the temperature was 27°C. We used QAOS (the Quantitative Amplification Of ssDNA) to detect ssDNA as in [29], [51]. (f–h) The dynamics of Sgs1 association with chromosome ends following telomere uncapping at 36°C, in cdc13-1 cells with a rif1Δ mutation (dark columns) or with wild-type RIF1 (light columns) at: (f) 1 kb, in sub-telomeres, (g) 8 kb and (h) 15 kb. Time 0 temperature and error bars are as in Figure 1. (i) A model of RPA and checkpoint proteins association with YER188W in rif1Δ (left) versus RIF1+ cells (right). Squares indicate chromosomal regions detected by ChIP and QAOS. (j–k) The ssDNA tolerance threshold model: (j) When short ssDNA lesions are generated, Rif1 acts as a “band-aid” for these lesions, associating with DNA-junctions and the adjacent ssDNA and blocking access for RPA and checkpoint proteins. In consequence, cells avoid spending energy to arrest and to re-start the cell cycle. (k) When longer ssDNA lesions are generated, they extend beyond Rif1-protected regions. In consequence, the damage is detected, a checkpoint signal is fired and cells arrest the cell cycle.