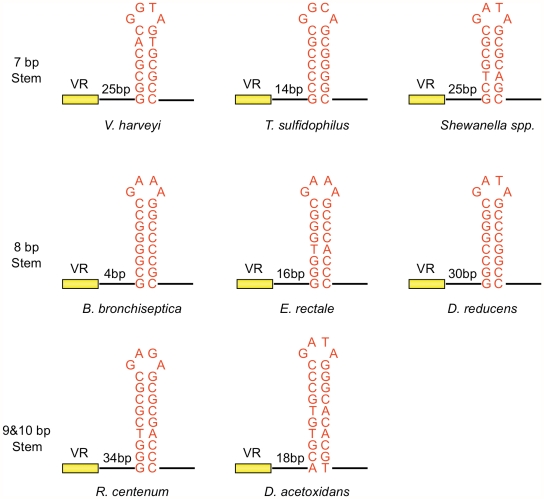
Figure 5. Phage-related DGRs contain potential hairpin/cruciform structures with conserved features.
DGRs associated with phages or phage-related sequences in different bacterial genomes were identified as described in Doulatov et al. [1]. Short inverted repeats which could potentially form hairpin/cruciform structures were found downstream of VRs as shown. In each case, GC-rich stems are 7–10 bp in length with 4 nt loops composed of the conserved sequence [5′GRNA; R = A or G, N = any nucleotide]. Relative distances between the hairpin/cruciform structures and their corresponding VRs range from 4 to 34 bp. V. harveyi, Vibrio harveyi phage VHML; T. sulfidophilus, Thioalkalivibrio sulfidophilus HL-EbGr7; Shewanella sp., Shewanella sp. W3-18-1; B. bronchiseptica, Bordetella bronchiseptica phage BPP-1; E. rectale, Eubacterium rectale DSM 17629; D. reducens, Desulfotomaculum reducens MI-1; R. centenum, Rhodospirillum centenum SW; D. acetoxidans, Desulfotomaculum acetoxidans DSM 771.