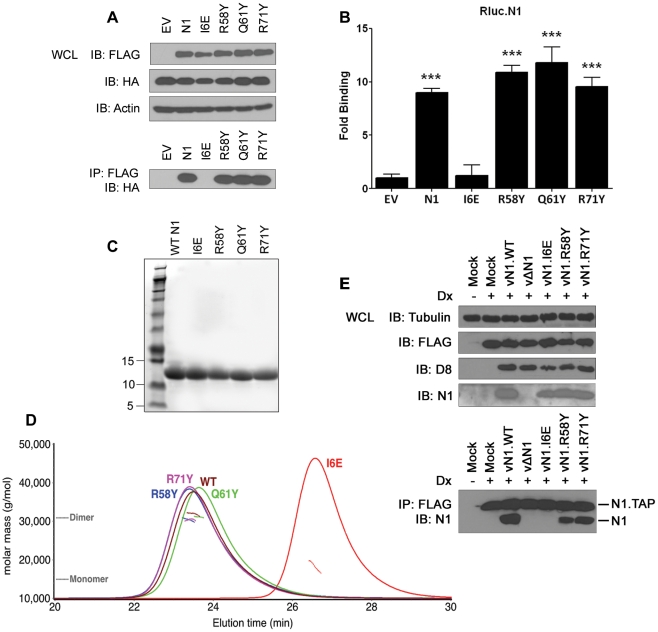
Figure 3. Biochemical and biophysical characterization of mutant N1.
(A) Ability of FLAG-tagged WT and mutant N1 to immunoprecipitate HA-tagged WT N1 after transfection in HEK 293T cells. (B) Ability of FLAG-tagged WT and mutant N1 to immunoprecipitate a renilla luciferase-fused WT N1 (Rluc.N1). Relative fold binding for each plasmid is calculated in triplicates after normalization to empty vector (EV). Data are expressed as means ± SD with statistical analysis (Student's t-test; ***P<0.0005). (C) Purification of WT and mutant N1 after over-expression in E. coli. (D) SEC-MALS curves obtained for WT and mutant N1. Weight-averaged molar mass (dotted lines) is shown across the elution profile (A280 nm, solid lines) of wild-type and mutant N1. While wild-type (WT) N1 and the groove-filling mutants elute as a dimer, I6E N1 is predominantly monomeric. (E) Ability of mutant N1 proteins expressed from recombinant VACV viruses to dimerise in HEK 293 T-REx cells with TAP-tagged N1 expressed after addition of doxycycline (Dx) for 2 h.