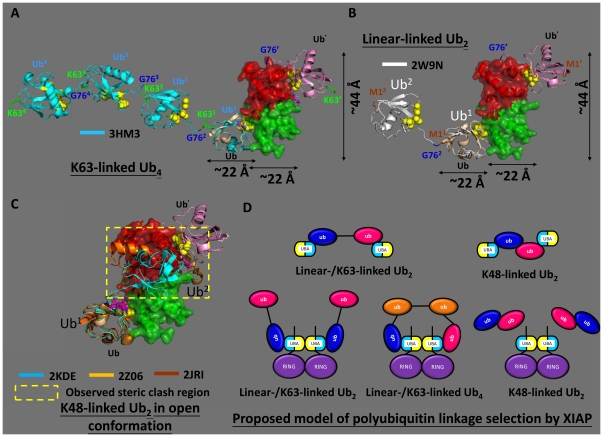
Figure 6. Superpositions of K48-, K63- and linear-linked polyubiquitins with the aligned model and the proposed mechanism of polyubiquitin linkage-selection by XIAP.
Superpositions of the most proximal unit Ub1 of K63-linked tetraubiquitin (A, cyan ribbon (3HM3)), linear-linked diubiquitin (B, white ribbon (2W9N)) and K48-linked diubiquitin (C, cyan (2KDE), orange (2Z06), brown (2JRI) ribbons) with the ribbon structure of Ub (wheat) on the aligned models from Figure 5G. The Ub′ (pink ribbon) indicates the ubiquitin which associated with the XIAP-UBA′ protomer. The superscript prime denotes residues from Ub′. The superscript numbering refers to the position of the ubiquitin units in the polyubiquitin chain such that the Ub1 and M11 denoted the most proximal unit and the Met1 of Ub1, respectively. The side-chains of residues M1 (brown), K63 (green) and G76 (blue) are labeled and shown in stick representation. The Leu8-Ile44-Val70-centred hydrophobic patch in ubiquitin is shown in yellow sphere. The XIAP-UBA and XIAP-UBA′ are shown in green and red semi-transparent surface. The region of steric clash between the Ub′ of the aligned models and the Ub2 of K48-linked diubiquitin is highlighted with a dotted rectangle in (C). (D) The speculative mechanism of polyubiquitin linkage-selection by XIAP. The XIAP-RING dimerization assists the oligomerization of XIAP-UBA, via the self-association (yellow) surface. The homodimeric XIAP-UBA arranges its ubiquitin-binding surfaces (cyan) into an orientation which can simultaneously recognize two separate (but not successive) ubiquitin units on the same flexible K63-/linear-linked polyubiquitin chain to achieve the high-affinity polyubiquitin chain binding (middle, bottom line). The binding of K48-linked polyubiquitin chain (in open conformation) with XIAP-UBA dimer is not spatially feasible (right, bottom line). Low-affinity binding events are observed between predominantly monomeric XIAP-UBA and the successive units of either K63-/linear-linked or K48-linked polyubiquitin. The observed subtle increase in binding affinity of diubiquitins to XIAP-UBA was probably due to increase of local concentration of ubiquitin in the diubiquitin chain (Top line). All figures were generated using PyMOL (DeLano Scientific).