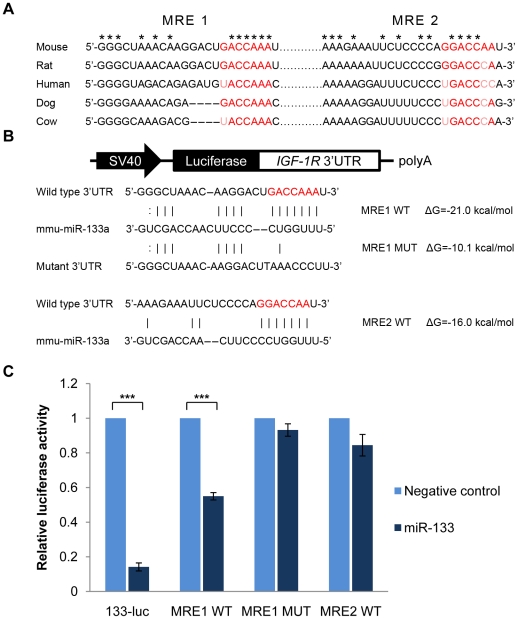
Figure 1. Identification of a functional miR-133 binding site in the IGF-1R 3′UTR.
(A) Seed-matched sequences in the IGF-1R 3′UTRs are in red and conserved regions between aligned sequences are indicated by stars. (B) Schematic representation of luciferase reporter constructs. miRNA-mRNA hybridization structures and folding energies were predicted by RNAhybrid. (C) HEK 293T cells were transfected with psiCHECK2 luciferase reporter vectors containing wild type or mutated miR-133 binding sites downstream of the Renilla luciferase gene (50 ng), and the internal Firefly luciferase gene was used to normalize for transfection efficiency. A pcDNA6.2-miR-133 expression vector or pcDNA6.2-negative control vector was cotransfected (150 ng). Dual-luciferase assays were performed 48 hours after transfection. Normalized luciferase activities of miR-133 transfectants were shown as the percentage relative to pcDNA6.2 transfectant, which was set at 1. Data represent the mean ± standard deviation (S.D.) of three independent experiments. ***p<0.001 vs. pcDNA6.2 transfectants.