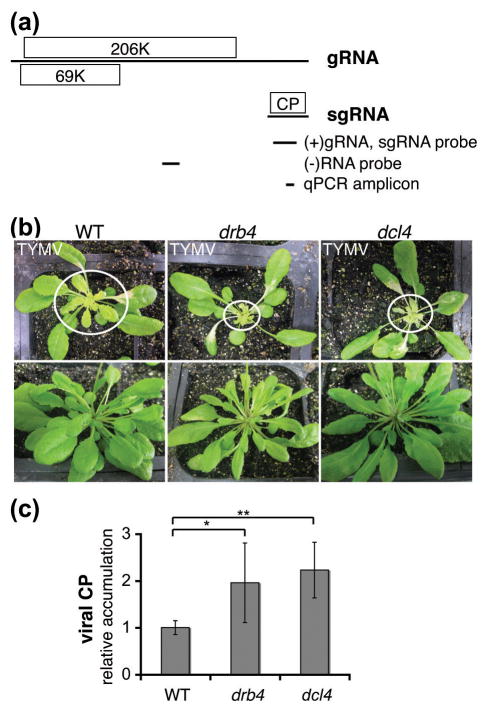
Figure 1. DRB4 and DCL4 contribute to antiviral response to TYMV.
(a) TYMV genome structure and expression.
TYMV genomic RNA (gRNA) directs the expression of a replication protein (206K) and an RNA silencing suppressor (69K). The subgenomic RNA (sgRNA), which is derived from the 3′ end of gRNA, directs the expression of the coat protein (CP).
A schematic representation of the Northern blot probes and qPCR amplicon used for viral RNA detection is shown below. Genomic coordinates are the following: 5717-6124nt for (+)gRNA and sgRNA detection, 3297-3558nt for (−)RNA detection and 6004-6066nt for qPCR amplicon.
(b) TYMV infection symptoms in WT and drb4 and dcl4 mutant plants.
Images of infected plants (upper panel) and healthy controls (lower panel) were taken 12 days post-infection. Systemically infected leaves are encircled. The enhanced infection symptoms in drb4 and dcl4 mutant plants were consistently observed in independent experiments (n ≥ 2).
(c) Accumulation of the TYMV CP is enhanced in drb4 and dcl4 mutant plants.
Extracts from systemically infected leaves of individual plants containing equal total protein amounts were subjected to ELISA using CP-specific antibody. The data is presented as a mean viral CP levels normalized by total protein content relative to the value obtained for WT plants. Error bars represent standard deviation (n=15–19 plants per genotype). The statistical significance was tested using two-tailed Mann-Whitney non-parametric test: * p value = 10−3, ** p value <10−5.