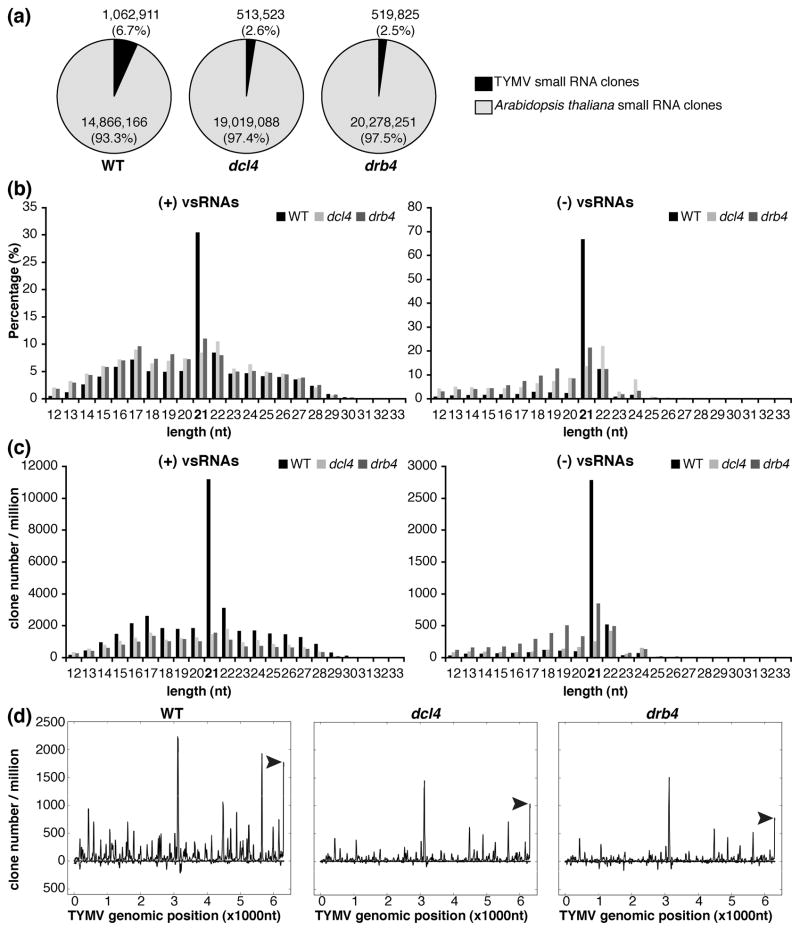
Figure 2. DCL4 and DRB4 are required for the biogenesis of vsRNAs.
RNA extracted from systemically infected leaves of WT and drb4 and dcl4 mutant plants were used to construct small RNA libraries for deep sequencing analyses.
(a) Virus- and host-specific small RNAs in WT, dcl4 mutant and drb4 mutant plants.
Sequence tags with a perfect match to the Arabidopsis genome are presented in light gray (14 866 166, 19 019 088 and 20 278 251 sRNA clones in the libraries obtained from WT, dcl4 and drb4 mutant plants, respectively). Sequence tags with a perfect match to TYMV genome are presented in black (1 062 911, 513 523 and 519 825 sRNA clones in the libraries obtained from WT, dcl4 and drb4 mutant plants, respectively). sRNA clones with a perfect match to both Arabidopsis and TYMV genomes (0.25%, 0.23% and 0.20% of mapped small RNAs in the libraries obtained from WT, dcl4 and drb4 mutant plants, respectively) are not presented.
(b, c) Length distribution of vsRNAs in WT, dcl4 mutant and drb4 mutant plants.
Size distribution of (+)vsRNAs (left panels) or (−)vsRNAs (right panels) is presented as the percentage of total (+)vsRNAs or (−)vsRNAs, respectively (b) and as the sum of library-size normalized sRNA counts (c) for each class.
(d) TYMV genomic view of vsRNAs in WT, dcl4 mutant and drb4 mutant plants.
(+)vsRNAs are presented above the X axis. (−)vsRNAs are shown below the X axis. The abundance of vsRNAs was calculated and plotted as the sum of library-size normalized sRNA counts in each single nucleotide sliding window along the TYMV genome. Peaks of vsRNAs derived from the tRNA-like structure at 3′ end of the genome are indicated by arrow heads.