Fig. 3.
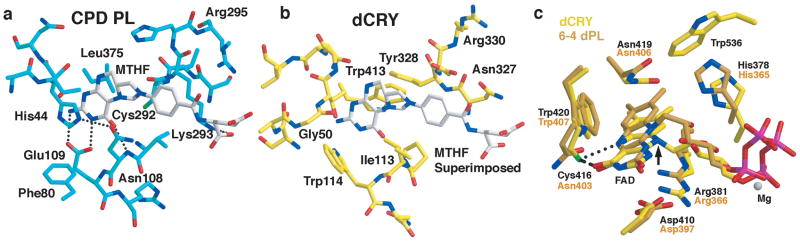
Cofactor binding regions of dCRY. a) E. coli CPD PL binds MTHF as an antenna cofactor. In dCRY (b), few of the residues that interact with MTFH in CPD PL are present, including the Glu residue that recognizes the pterin ring or otherwise contact MTHF (E. coli PL Glu109 → dCRY Trp114, Asn108 → Ile113, His44 → Gly50). Loop regions surrounding the cofactor in dCRY and 6-4 dPL also have different structures and compositions (e.g. 6-4 dPL Ile50 → dCRY Glu45, Leu51 → Ser46, Leu40 → Phe42, Met54 → Gly50) c) Flavin center of dCRY (yellow) compared to 6-4 PL (orange). dCRY Cys416 replaces 6-4 PL Asn and is in hydrogen bonding distance (dotted lines) of both N5 and O4 of FAD. The linkage between the ring and ribose is altered in dCRY by semiquinone formation.